Sexual Differentiation Is Coordinately Regulated by Cryptococcus neoformans CRK1 and GAT1
- PMID: 32575488
- PMCID: PMC7349709
- DOI: 10.3390/genes11060669
Sexual Differentiation Is Coordinately Regulated by Cryptococcus neoformans CRK1 and GAT1
Abstract
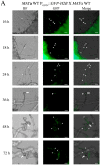
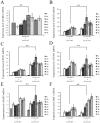
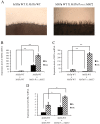
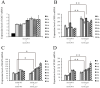
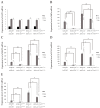
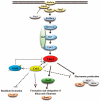
The heterothallic basidiomycetous fungus Cryptococcus neoformans has two mating types, MATa and MATα. Morphological progression of bisexual reproduction in C. neoformans is as follows: yeast to hyphal transition, filament extension, basidium formation, meiosis, and sporulation. C. neoformans Cdk-related kinase 1 (CRK1) is a negative regulator of bisexual mating. In this study, we characterized the morphological features of mating structures in the crk1 mutant and determined the genetic interaction of CRK1 in the regulatory networks of sexual differentiation. In the bilateral crk1 mutant cross, despite shorter length of filaments than in the wild-type cross, dikaryotic filaments and other structures still remained intact during bisexual mating, but the timing of basidium formation was approximately 18 h earlier than in the cross between wild type strains. Furthermore, gene expression analyses revealed that CRK1 modulated the expression of genes involved in the progression of hyphal elongation, basidium formation, karyogamy and meiosis. Phenotypic results showed that, although deletion of C. neoformans CRK1 gene increased the efficiency of bisexual mating, filamentation in the crk1 mutant was blocked by MAT2 or ZNF2 mutation. A bioinformatics survey predicted the C. neoformans GATA transcriptional factor Gat1 as a potential substrate of Crk1 kinase. Our genetic and phenotypic findings revealed that C. neoformansGAT1 and CRK1 formed a regulatory circuit to negatively regulate MAT2 to control filamentation progression and transition during bisexual mating.
Keywords: CRK1; Cryptococcus neoformans; GAT1; bisexual mating; filament differentiation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






Similar articles
-
Mating differentiation in Cryptococcus neoformans is negatively regulated by the Crk1 protein kinase.Fungal Genet Biol. 2011 Mar;48(3):225-40. doi: 10.1016/j.fgb.2010.11.005. Epub 2010 Nov 24. Fungal Genet Biol. 2011. PMID: 21111055
-
PRM1 and KAR5 function in cell-cell fusion and karyogamy to drive distinct bisexual and unisexual cycles in the Cryptococcus pathogenic species complex.PLoS Genet. 2017 Nov 27;13(11):e1007113. doi: 10.1371/journal.pgen.1007113. eCollection 2017 Nov. PLoS Genet. 2017. PMID: 29176784 Free PMC article.
-
Uniparental nuclear inheritance following bisexual mating in fungi.Elife. 2021 Aug 2;10:e66234. doi: 10.7554/eLife.66234. Elife. 2021. PMID: 34338631 Free PMC article.
-
Unisexual versus bisexual mating in Cryptococcus neoformans: Consequences and biological impacts.Fungal Genet Biol. 2015 May;78:65-75. doi: 10.1016/j.fgb.2014.08.008. Epub 2014 Aug 27. Fungal Genet Biol. 2015. PMID: 25173822 Free PMC article. Review.
-
The role of mating type and morphology in Cryptococcus neoformans pathogenesis.Int J Med Microbiol. 2002 Oct;292(5-6):313-29. doi: 10.1078/1438-4221-00216. Int J Med Microbiol. 2002. PMID: 12452279 Review.
Cited by
-
A Limited Number of Amino Acid Permeases Are Crucial for Cryptococcus neoformans Survival and Virulence.Int J Microbiol. 2024 Aug 8;2024:5566438. doi: 10.1155/2024/5566438. eCollection 2024. Int J Microbiol. 2024. PMID: 39148675 Free PMC article. Review.
-
Ubiquitin proteolysis of a CDK-related kinase regulates titan cell formation and virulence in the fungal pathogen Cryptococcus neoformans.Nat Commun. 2022 Oct 27;13(1):6397. doi: 10.1038/s41467-022-34151-6. Nat Commun. 2022. PMID: 36302775 Free PMC article.
-
Is Cryptococcus neoformans a pleomorphic fungus?Curr Opin Microbiol. 2024 Dec;82:102539. doi: 10.1016/j.mib.2024.102539. Epub 2024 Sep 10. Curr Opin Microbiol. 2024. PMID: 39260180 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

