Optimized qRT-PCR Approach for the Detection of Intra- and Extra-Cellular SARS-CoV-2 RNAs
- PMID: 32575728
- PMCID: PMC7352576
- DOI: 10.3390/ijms21124396
Optimized qRT-PCR Approach for the Detection of Intra- and Extra-Cellular SARS-CoV-2 RNAs
Abstract
The novel coronavirus SARS-CoV-2 is the causative agent of the acute respiratory disease COVID-19, which has become a global concern due to its rapid spread. Meanwhile, increased demand for testing has led to a shortage of reagents and supplies and compromised the performance of diagnostic laboratories in many countries. Both the World Health Organization (WHO) and the Center for Disease Control and Prevention (CDC) recommend multi-step RT-PCR assays using multiple primer and probe pairs, which might complicate the interpretation of the test results, especially for borderline cases. In this study, we describe an alternative RT-PCR approach for the detection of SARS-CoV-2 RNA that can be used for the probe-based detection of clinical isolates in diagnostics as well as in research labs using a low-cost SYBR green method. For the evaluation, we used samples from patients with confirmed SARS-CoV-2 infections and performed RT-PCR assays along with successive dilutions of RNA standards to determine the limit of detection. We identified an M-gene binding primer and probe pair highly suitable for the quantitative detection of SARS-CoV-2 RNA for diagnostic and research purposes.
Keywords: COVID-19; SARS-CoV-2; coronavirus; qRT-PCR detection; test protocol.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

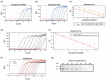

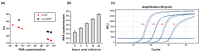

References
-
- WHO . WHO SARS-CoV-2 Situation Report. WHO; Geneva, Switzerland: 2020.
-
- Centers for Disease Control and Prevention CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. Revision. 2020;3:30.
-
- Drosten C., Günther S., Preiser W., Van Der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R., et al. Identification of a Novel Coronavirus in Patients with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

