Myoferlin Is a Yet Unknown Interactor of the Mitochondrial Dynamics' Machinery in Pancreas Cancer Cells
- PMID: 32575867
- PMCID: PMC7352660
- DOI: 10.3390/cancers12061643
Myoferlin Is a Yet Unknown Interactor of the Mitochondrial Dynamics' Machinery in Pancreas Cancer Cells
Abstract
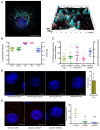
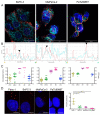
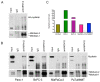
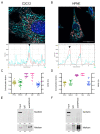
Pancreas ductal adenocarcinoma is one of the deadliest cancers where surgery remains the main survival factor. Mitochondria were described to be involved in tumor aggressiveness in several cancer types including pancreas cancer. We have previously reported that myoferlin controls mitochondrial structure and function, and demonstrated that myoferlin depletion disturbs the mitochondrial dynamics culminating in a mitochondrial fission. In order to unravel the mechanism underlying this observation, we explored the myoferlin localization in pancreatic cancer cells and showed a colocalization with the mitochondrial dynamic machinery element: mitofusin. This colocalization was confirmed in several pancreas cancer cell lines and in normal cell lines as well. Moreover, in pancreas cancer cell lines, it appeared that myoferlin interacted with mitofusin. These discoveries open-up new research avenues aiming at modulating mitofusin function in pancreas cancer.
Keywords: mitochondria; mitofusin; myoferlin; pancreas cancer.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Rademaker G., Costanza B., Anania S., Agirman F., Maloujahmoum N., Valentin E.D., Goval J.J., Bellahcène A., Castronovo V., Peulen O.J. Myoferlin Contributes to the Metastatic Phenotype of Pancreatic Cancer Cells by Enhancing Their Migratory Capacity through the Control of Oxidative Phosphorylation. Cancers. 2019;11:853. doi: 10.3390/cancers11060853. - DOI - PMC - PubMed
-
- Rademaker G., Hennequière V., Brohée L., Nokin M.-J., Lovinfosse P., Durieux F., Gofflot S., Bellier J., Costanza B., Herfs M., et al. Myoferlin controls mitochondrial structure and activity in pancreatic ductal adenocarcinoma, and affects tumor aggressiveness. Oncogene. 2018;66:1–15. doi: 10.1038/s41388-018-0287-z. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

