A ligation and restriction enzyme independent cloning technique: an alternative to conventional methods for cloning hard-to-clone gene segments in the influenza reverse genetics system
- PMID: 32576218
- PMCID: PMC7309217
- DOI: 10.1186/s12985-020-01358-2
A ligation and restriction enzyme independent cloning technique: an alternative to conventional methods for cloning hard-to-clone gene segments in the influenza reverse genetics system
Abstract
Background: Reverse genetics is used in many laboratories around the world and enables the creation of tailor-made influenza viruses with a desired genotype or phenotype. However, the process is not flawless, and difficulties remain during cloning of influenza gene segments into reverse genetics vectors (pHW2000, pHH21, pCAGGS). Reverse genetics begins with making cDNA copies of influenza gene segments and cloning them into bi-directional (pHW2000) or uni-directional plasmids (pHH21, pCAGGS) followed by transfection of the recombinant plasmid(s) to HEK-293 T or any other suitable cells which are permissive to transfection. However, the presence of internal restriction sites in the gene segments of many field isolates of avian influenza viruses makes the cloning process difficult, if employing conventional methods. Further, the genetic instability of influenza gene-containing plasmids in bacteria (especially Polymerase Basic 2 and Polymerase Basic 1 genes; PB2 and PB1) also leads to erroneous incorporation of bacterial genomic sequences into the influenza gene of interest.
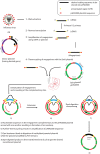
Methods: Herein, we report an easy and efficient ligation and restriction enzyme independent (LREI) cloning method for cloning influenza gene segments into pHW2000 vector. The method involves amplification of megaprimers followed by PCR amplification of megaprimers using a bait plasmid, DpnI digestion and transformation.
Results: Hard-to-clone genes: PB2 of A/chicken/Bangladesh/23527/2014 (H9N2) and PB1 of A/chicken/Bangladesh/23527/2014 (H9N2), A/chicken/Jiangxi/02.05YGYXG023-P/2015 (H5N6) and A/Chicken/Vietnam/H7F-14-BN4-315/2014 (H9N2) were cloned into pHW2000 using our LREI method and recombinant viruses were subsequently rescued.
Conclusion: The LREI cloning procedure represents an alternative strategy for cloning influenza gene segments which have internal restriction sites for the enzymes used in reverse genetics. Further, the problem of genetic instability in bacteria can be alleviated by growing recombinant bacterial cultures at a lower temperature. This technique can be applied to clone any influenza gene segment using universal primers, which would help in rapid generation of influenza viruses and facilitate influenza research and vaccine development.
Keywords: Influenza; Polymerase; Restriction enzyme independent cloning; Reverse genetics.
Conflict of interest statement
The authors declare that they have no competing interests. The funders had no role in the design of the study; in the writing of the manuscript, or in the decision to publish the results.
Figures


Similar articles
-
Development of a rapid, simple and efficient one-pot cloning method for a reverse genetics system of broad subtypes of influenza A virus.Sci Rep. 2019 Jun 5;9(1):8318. doi: 10.1038/s41598-019-44813-z. Sci Rep. 2019. PMID: 31165766 Free PMC article.
-
An efficient and rapid influenza gene cloning strategy for reverse genetics system.J Virol Methods. 2015 Sep 15;222:91-4. doi: 10.1016/j.jviromet.2015.06.001. Epub 2015 Jun 6. J Virol Methods. 2015. PMID: 26057220
-
Rapid and reliable universal cloning of influenza A virus genes by target-primed plasmid amplification.Nucleic Acids Res. 2008 Dec;36(21):e139. doi: 10.1093/nar/gkn646. Epub 2008 Oct 2. Nucleic Acids Res. 2008. PMID: 18832366 Free PMC article.
-
Reverse genetics plasmid for cloning unstable influenza A virus gene segments.J Virol Methods. 2011 May;173(2):378-83. doi: 10.1016/j.jviromet.2011.01.021. Epub 2011 Feb 3. J Virol Methods. 2011. PMID: 21295611 Free PMC article.
-
Engineering influenza viral vectors.Bioengineered. 2013 Jan-Feb;4(1):9-14. doi: 10.4161/bioe.21950. Epub 2012 Aug 24. Bioengineered. 2013. PMID: 22922205 Free PMC article. Review.
Cited by
-
Coinfection of Chickens with H9N2 and H7N9 Avian Influenza Viruses Leads to Emergence of Reassortant H9N9 Virus with Increased Fitness for Poultry and a Zoonotic Potential.J Virol. 2022 Mar 9;96(5):e0185621. doi: 10.1128/jvi.01856-21. Epub 2022 Jan 12. J Virol. 2022. PMID: 35019727 Free PMC article.
-
Silencing Transcription from an Influenza Reverse Genetics Plasmid in E. coli Enhances Gene Stability.ACS Synth Biol. 2023 Feb 17;12(2):432-445. doi: 10.1021/acssynbio.2c00358. Epub 2023 Jan 30. ACS Synth Biol. 2023. PMID: 36716395 Free PMC article.
-
The Origin of Internal Genes Contributes to the Replication and Transmission Fitness of H7N9 Avian Influenza Virus.J Virol. 2022 Nov 23;96(22):e0129022. doi: 10.1128/jvi.01290-22. Epub 2022 Nov 7. J Virol. 2022. PMID: 36342296 Free PMC article.
-
Membrane Permeabilization and Antimicrobial Activity of Recombinant Defensin-d2 and Actifensin against Multidrug-Resistant Pseudomonas aeruginosa and Candida albicans.Molecules. 2022 Jul 6;27(14):4325. doi: 10.3390/molecules27144325. Molecules. 2022. PMID: 35889198 Free PMC article.
-
Research of recombinant influenza A virus as a vector for Mycoplasma pneumoniae P1a and P30a.Immun Inflamm Dis. 2024 Sep;12(9):e70021. doi: 10.1002/iid3.70021. Immun Inflamm Dis. 2024. PMID: 39291404 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- BB/R012679/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/I/00007034/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/S013792/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/N002571/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/I/00007035/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

