EBV renders B cells susceptible to HIV-1 in humanized mice
- PMID: 32576602
- PMCID: PMC7335381
- DOI: 10.26508/lsa.202000640
EBV renders B cells susceptible to HIV-1 in humanized mice
Abstract
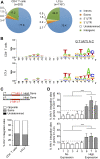
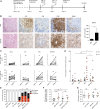
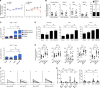
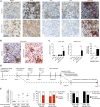
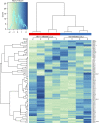
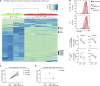
HIV and EBV are human pathogens that cause a considerable burden to worldwide health. In combination, these viruses are linked to AIDS-associated lymphomas. We found that EBV, which transforms B cells, renders them susceptible to HIV-1 infection in a CXCR4 and CD4-dependent manner in vitro and that CXCR4-tropic HIV-1 integrates into the genome of these B cells with the same molecular profile as in autologous CD4+ T cells. In addition, we established a humanized mouse model to investigate the in vivo interactions of EBV and HIV-1 upon coinfection. The respective mice that reconstitute human immune system components upon transplantation with CD34+ human hematopoietic progenitor cells could recapitulate aspects of EBV and HIV immunobiology observed in dual-infected patients. Upon coinfection of humanized mice, EBV/HIV dual-infected B cells could be detected, but were susceptible to CD8+ T-cell-mediated immune control.
© 2020 McHugh et al.
Conflict of interest statement
RF Speck and R Myburgh are shareholders of Transcure Bioservices (
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
