GapMind: Automated Annotation of Amino Acid Biosynthesis
- PMID: 32576650
- PMCID: PMC7311316
- DOI: 10.1128/mSystems.00291-20
GapMind: Automated Annotation of Amino Acid Biosynthesis
Abstract
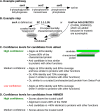
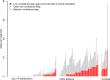
GapMind is a Web-based tool for annotating amino acid biosynthesis in bacteria and archaea (http://papers.genomics.lbl.gov/gaps). GapMind incorporates many variant pathways and 130 different reactions, and it analyzes a genome in just 15 s. To avoid error-prone transitive annotations, GapMind relies primarily on a database of experimentally characterized proteins. GapMind correctly handles fusion proteins and split proteins, which often cause errors for best-hit approaches. To improve GapMind's coverage, we examined genetic data from 35 bacteria that grow in defined media without amino acids, and we filled many gaps in amino acid biosynthesis pathways. For example, we identified additional genes for arginine synthesis with succinylated intermediates in Bacteroides thetaiotaomicron, and we propose that Dyella japonica synthesizes tyrosine from phenylalanine. Nevertheless, for many bacteria and archaea that grow in minimal media, genes for some steps still cannot be identified. To help interpret potential gaps, GapMind checks if they match known gaps in related microbes that can grow in minimal media. GapMind should aid the identification of microbial growth requirements.IMPORTANCE Many microbes can make all of the amino acids (the building blocks of proteins). In principle, we should be able to predict which amino acids a microbe can make, and which it requires as nutrients, by checking its genome sequence for all of the necessary genes. However, in practice, it is difficult to check for all of the alternative pathways. Furthermore, new pathways and enzymes are still being discovered. We built an automated tool, GapMind, to annotate amino acid biosynthesis in bacterial and archaeal genomes. We used GapMind to list gaps: cases where a microbe makes an amino acid but a complete pathway cannot be identified in its genome. We used these gaps, together with data from mutants, to identify new pathways and enzymes. However, for most bacteria and archaea, we still do not know how they can make all of the amino acids.
Keywords: amino acid biosynthesis; gene annotation; high-throughput genetics.
Copyright © 2020 Price et al.
Figures




Similar articles
-
Filling gaps in bacterial catabolic pathways with computation and high-throughput genetics.PLoS Genet. 2022 Apr 13;18(4):e1010156. doi: 10.1371/journal.pgen.1010156. eCollection 2022 Apr. PLoS Genet. 2022. PMID: 35417463 Free PMC article.
-
Filling gaps in bacterial amino acid biosynthesis pathways with high-throughput genetics.PLoS Genet. 2018 Jan 11;14(1):e1007147. doi: 10.1371/journal.pgen.1007147. eCollection 2018 Jan. PLoS Genet. 2018. PMID: 29324779 Free PMC article.
-
[Analysis, identification and correction of some errors of model refseqs appeared in NCBI Human Gene Database by in silico cloning and experimental verification of novel human genes].Yi Chuan Xue Bao. 2004 May;31(5):431-43. Yi Chuan Xue Bao. 2004. PMID: 15478601 Chinese.
-
Variations in metabolic pathways create challenges for automated metabolic reconstructions: Examples from the tetrahydrofolate synthesis pathway.Comput Struct Biotechnol J. 2014 Jun 11;10(16):41-50. doi: 10.1016/j.csbj.2014.05.008. eCollection 2014 Jun. Comput Struct Biotechnol J. 2014. PMID: 25210598 Free PMC article. Review.
-
Genomic attributes of thermophilic and hyperthermophilic bacteria and archaea.World J Microbiol Biotechnol. 2022 Jun 13;38(8):135. doi: 10.1007/s11274-022-03327-z. World J Microbiol Biotechnol. 2022. PMID: 35695998 Review.
Cited by
-
Reduced and Nonreduced Genomes in Paraburkholderia Symbionts of Social Amoebas.mSystems. 2022 Oct 26;7(5):e0056222. doi: 10.1128/msystems.00562-22. Epub 2022 Sep 13. mSystems. 2022. PMID: 36098425 Free PMC article.
-
Genetic Engineering of Resident Bacteria in the Gut Microbiome.J Bacteriol. 2023 Jul 25;205(7):e0012723. doi: 10.1128/jb.00127-23. Epub 2023 Jun 29. J Bacteriol. 2023. PMID: 37382533 Free PMC article. Review.
-
Leveraging genomic information to predict environmental preferences of bacteria.ISME J. 2024 Jan 8;18(1):wrae195. doi: 10.1093/ismejo/wrae195. ISME J. 2024. PMID: 39361898 Free PMC article. Review.
-
Integrating biological knowledge for mechanistic inference in the host-associated microbiome.Front Microbiol. 2024 Apr 4;15:1351678. doi: 10.3389/fmicb.2024.1351678. eCollection 2024. Front Microbiol. 2024. PMID: 38638909 Free PMC article. Review.
-
Comparative analysis of amino acid auxotrophies and peptidase profiles in non-dysbiotic and dysbiotic small intestinal microbiomes.Comput Struct Biotechnol J. 2025 Feb 12;27:821-831. doi: 10.1016/j.csbj.2025.02.004. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40103612 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
