High-Resolution Longitudinal Dynamics of the Cystic Fibrosis Sputum Microbiome and Metabolome through Antibiotic Therapy
- PMID: 32576651
- PMCID: PMC7311317
- DOI: 10.1128/mSystems.00292-20
High-Resolution Longitudinal Dynamics of the Cystic Fibrosis Sputum Microbiome and Metabolome through Antibiotic Therapy
Abstract
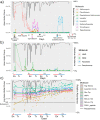
Microbial diversity in the cystic fibrosis (CF) lung decreases over decades as pathogenic bacteria such as Pseudomonas aeruginosa take over. The dynamics of the CF microbiome and metabolome over shorter time frames, however, remain poorly studied. Here, we analyze paired microbiome and metabolome data from 594 sputum samples collected over 401 days from six adult CF subjects (subject mean = 179 days) through periods of clinical stability and 11 CF pulmonary exacerbations (CFPE). While microbiome profiles were personalized (permutational multivariate analysis of variance [PERMANOVA] r 2 = 0.79, P < 0.001), we observed significant intraindividual temporal variation that was highest during clinical stability (linear mixed-effects [LME] model, P = 0.002). This included periods where the microbiomes of different subjects became highly similar (UniFrac distance, <0.05). There was a linear increase in the microbiome alpha-diversity and in the log ratio of anaerobes to pathogens with time (n = 14 days) during the development of a CFPE (LME P = 0.0045 and P = 0.029, respectively). Collectively, comparing samples across disease states showed there was a reduction of these two measures during antibiotic treatment (LME P = 0.0096 and P = 0.014, respectively), but the stability data and CFPE data were not significantly different from each other. Metabolome alpha-diversity was higher during CFPE than during stability (LME P = 0.0085), but no consistent metabolite signatures of CFPE across subjects were identified. Virulence-associated metabolites from P. aeruginosa were temporally dynamic but were not associated with any disease state. One subject died during the collection period, enabling a detailed look at changes in the 194 days prior to death. This subject had over 90% Pseudomonas in the microbiome at the beginning of sampling, and that level gradually increased to over 99% prior to death. This study revealed that the CF microbiome and metabolome of some subjects are dynamic through time. Future work is needed to understand what drives these temporal dynamics and if reduction of anaerobes correlate to clinical response to CFPE therapy.IMPORTANCE Subjects with cystic fibrosis battle polymicrobial lung infections throughout their lifetime. Although antibiotic therapy is a principal treatment for CF lung disease, we have little understanding of how antibiotics affect the CF lung microbiome and metabolome and how much the community changes on daily timescales. By analyzing 594 longitudinal CF sputum samples from six adult subjects, we show that the sputum microbiome and metabolome are dynamic. Significant changes occur during times of stability and also through pulmonary exacerbations (CFPEs). Microbiome alpha-diversity increased as a CFPE developed and then decreased during treatment in a manner corresponding to the reduction in the log ratio of anaerobic bacteria to classic pathogens. Levels of metabolites from the pathogen P. aeruginosa were also highly variable through time and were negatively associated with anaerobes. The microbial dynamics observed in this study may have a significant impact on the outcome of antibiotic therapy for CFPEs and overall subject health.
Keywords: antibiotics; cystic fibrosis; metabolome; microbiome.
Copyright © 2020 Raghuvanshi et al.
Figures


References
-
- Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, Cavalcoli JD, VanDevanter DR, Murray S, Li JZ, Young VB, LiPuma JJ. 2012. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci U S A 109:5809–5814. doi:10.1073/pnas.1120577109. - DOI - PMC - PubMed
-
- Quinn RA, Whiteson K, Lim Y-W, Salamon P, Bailey B, Mienardi S, Sanchez SE, Blake D, Conrad D, Rohwer F. 2015. A Winogradsky-based culture system shows an association between microbial fermentation and cystic fibrosis exacerbation. ISME J 9:1052–1052. doi:10.1038/ismej.2014.266. - DOI - PMC - PubMed
