Genotypic and Phenotypic Diversity of Staphylococcus aureus Isolates from Cystic Fibrosis Patient Lung Infections and Their Interactions with Pseudomonas aeruginosa
- PMID: 32576671
- PMCID: PMC7315118
- DOI: 10.1128/mBio.00735-20
Genotypic and Phenotypic Diversity of Staphylococcus aureus Isolates from Cystic Fibrosis Patient Lung Infections and Their Interactions with Pseudomonas aeruginosa
Abstract
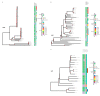
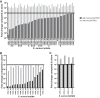
Staphylococcus aureus has recently overtaken Pseudomonas aeruginosa as the most commonly recognized bacterial pathogen that infects the respiratory tracts of individuals with the genetic disease cystic fibrosis (CF) in the United States. Most studies of S. aureus in CF patient lung infections have focused on a few isolates, often exclusively laboratory-adapted strains, and how they are killed by P. aeruginosa Less is known about the diversity of S. aureus CF patient lung isolates in terms of both their virulence and their interaction with P. aeruginosa To begin to address this gap, we recently sequenced 64 clinical S. aureus isolates and a reference isolate, JE2. Here, we analyzed the antibiotic resistance genotypes, sequence types, clonal complexes, spa types, agr types, and presence/absence of other known virulence factor genes of these isolates. We hypothesized that virulence phenotypes of S. aureus, namely, toxin production and the mucoid phenotype, would be lost in these isolates due to adaptation in the CF patient lung. In contrast to these expectations, we found that most isolates can lyse both rabbit and sheep blood (67.7%) and produce polysaccharide (69.2%), suggesting that these phenotypes were not lost during adaptation to the CF lung. We also identified three distinct phenotypic groups of S. aureus based on their survival in the presence of nonmucoid P. aeruginosa laboratory strain PAO1 and its mucoid derivative. Altogether, our work provides greater insight into the diversity of S. aureus isolates from CF patients, specifically the distribution of important virulence factors and their interaction with P. aeruginosa, all of which have implications in patient health.IMPORTANCEStaphylococcus aureus is now the most frequently detected recognized pathogen in the lungs of individuals who have cystic fibrosis (CF) in the United States, followed closely by Pseudomonas aeruginosa When these pathogens are found to coinfect the CF lung, patients have a significantly worse prognosis. While P. aeruginosa has been rigorously studied in the context of bacterial pathogenesis in CF, less is known about S. aureus Here, we present an in-depth study of 64 S. aureus clinical isolates from CF patients, for which we investigated genetic diversity utilizing whole-genome sequencing, virulence phenotypes, and interactions with P. aeruginosa We found that S. aureus isolated from CF lungs are phylogenetically diverse; most retain known virulence factors and vary in their interactions with P. aeruginosa (i.e., they range from being highly sensitive to P. aeruginosa to completely tolerant to it). Deepening our understanding of how S. aureus responds to its environment and other microbes in the CF lung will enable future development of effective treatments and preventative measures against these formidable infections.
Keywords: Pseudomonas aeruginosa; Staphylococcus aureus; cystic fibrosis; interspecies competition; phylogenetic analysis.
Copyright © 2020 Bernardy et al.
Figures

Similar articles
-
Association of Diverse Staphylococcus aureus Populations with Pseudomonas aeruginosa Coinfection and Inflammation in Cystic Fibrosis Airway Infection.mSphere. 2021 Jun 30;6(3):e0035821. doi: 10.1128/mSphere.00358-21. Epub 2021 Jun 23. mSphere. 2021. PMID: 34160233 Free PMC article.
-
Staphylococcus aureus and Pseudomonas aeruginosa Isolates from the Same Cystic Fibrosis Respiratory Sample Coexist in Coculture.Microbiol Spectr. 2022 Aug 31;10(4):e0097622. doi: 10.1128/spectrum.00976-22. Epub 2022 Jul 18. Microbiol Spectr. 2022. PMID: 35867391 Free PMC article.
-
Exogenous Alginate Protects Staphylococcus aureus from Killing by Pseudomonas aeruginosa.J Bacteriol. 2020 Mar 26;202(8):e00559-19. doi: 10.1128/JB.00559-19. Print 2020 Mar 26. J Bacteriol. 2020. PMID: 31792010 Free PMC article.
-
Help, hinder, hide and harm: what can we learn from the interactions between Pseudomonas aeruginosa and Staphylococcus aureus during respiratory infections?Thorax. 2019 Jul;74(7):684-692. doi: 10.1136/thoraxjnl-2018-212616. Epub 2019 Feb 18. Thorax. 2019. PMID: 30777898 Free PMC article. Review.
-
The Yin and Yang of Streptococcus Lung Infections in Cystic Fibrosis: a Model for Studying Polymicrobial Interactions.J Bacteriol. 2019 May 8;201(11):e00115-19. doi: 10.1128/JB.00115-19. Print 2019 Jun 1. J Bacteriol. 2019. PMID: 30885933 Free PMC article. Review.
Cited by
-
The impact of CFTR modulator triple therapy on type 2 inflammatory response in patients with cystic fibrosis.Allergy Asthma Clin Immunol. 2023 Jul 31;19(1):66. doi: 10.1186/s13223-023-00822-2. Allergy Asthma Clin Immunol. 2023. PMID: 37525180 Free PMC article.
-
Bacterial evolution during human infection: Adapt and live or adapt and die.PLoS Pathog. 2021 Sep 9;17(9):e1009872. doi: 10.1371/journal.ppat.1009872. eCollection 2021 Sep. PLoS Pathog. 2021. PMID: 34499699 Free PMC article. Review.
-
Epidemiology and impact of methicillin-sensitive Staphylococcus aureus with β-lactam antibiotic inoculum effects in adults with cystic fibrosis.Antimicrob Agents Chemother. 2023 Dec 14;67(12):e0013623. doi: 10.1128/aac.00136-23. Epub 2023 Nov 15. Antimicrob Agents Chemother. 2023. PMID: 37966229 Free PMC article.
-
Short chain fatty acids reduce the respiratory burst of human neutrophils in response to cystic fibrosis isolates of Staphylococcus aureus.J Cyst Fibros. 2023 Jul;22(4):756-762. doi: 10.1016/j.jcf.2023.04.022. Epub 2023 May 19. J Cyst Fibros. 2023. PMID: 37211502 Free PMC article.
-
Polymicrobial Interactions in the Cystic Fibrosis Airway Microbiome Impact the Antimicrobial Susceptibility of Pseudomonas aeruginosa.Antibiotics (Basel). 2021 Jul 7;10(7):827. doi: 10.3390/antibiotics10070827. Antibiotics (Basel). 2021. PMID: 34356747 Free PMC article. Review.
References
-
- Cystic Fibrosis Foundation. 2018. Cystic Fibrosis Foundation patient registry 2018 annual data report. Cystic Fibrosis Foundation, Bethesda, MD.
-
- Cystic Fibrosis Trust. 2009. Antibiotic treatment for cystic fibrosis—report of the UK Cystic Fibrosis Trust Antibiotic Working Group. Cystic Fibrosis Trust, London, United Kingdom.
-
- Mogayzel PJ, Naureckas ET, Robinson KA, Mueller G, Hadjiliadis D, Hoag JB, Lubsch L, Hazle L, Sabadosa K, Marshall B, Pulmonary Clinical Practice Guidelines Committee. 2013. Cystic fibrosis pulmonary guidelines. Chronic medications for maintenance of lung health. Am J Respir Crit Care Med 187:680–689. doi:10.1164/rccm.201207-1160oe. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

