HGT in the human and skin commensal Malassezia: A bacterially derived flavohemoglobin is required for NO resistance and host interaction
- PMID: 32576698
- PMCID: PMC7354939
- DOI: 10.1073/pnas.2003473117
HGT in the human and skin commensal Malassezia: A bacterially derived flavohemoglobin is required for NO resistance and host interaction
Abstract
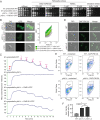
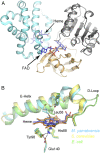
The skin of humans and animals is colonized by commensal and pathogenic fungi and bacteria that share this ecological niche and have established microbial interactions. Malassezia are the most abundant fungal skin inhabitant of warm-blooded animals and have been implicated in skin diseases and systemic disorders, including Crohn's disease and pancreatic cancer. Flavohemoglobin is a key enzyme involved in microbial nitrosative stress resistance and nitric oxide degradation. Comparative genomics and phylogenetic analyses within the Malassezia genus revealed that flavohemoglobin-encoding genes were acquired through independent horizontal gene transfer events from different donor bacteria that are part of the mammalian microbiome. Through targeted gene deletion and functional complementation in Malassezia sympodialis, we demonstrated that bacterially derived flavohemoglobins are cytoplasmic proteins required for nitric oxide detoxification and nitrosative stress resistance under aerobic conditions. RNA-sequencing analysis revealed that endogenous accumulation of nitric oxide resulted in up-regulation of genes involved in stress response and down-regulation of the MalaS7 allergen-encoding genes. Solution of the high-resolution X-ray crystal structure of Malassezia flavohemoglobin revealed features conserved with both bacterial and fungal flavohemoglobins. In vivo pathogenesis is independent of Malassezia flavohemoglobin. Lastly, we identified an additional 30 genus- and species-specific horizontal gene transfer candidates that might have contributed to the evolution of this genus as the most common inhabitants of animal skin.
Keywords: Malassezia; flavohemoglobin; horizontal gene transfer.
Conflict of interest statement
The authors declare no competing interest.
Figures




References
-
- Theelen B., et al. ., Malassezia ecology, pathophysiology, and treatment. Med. Mycol. 56 (suppl. 1), S10–S25 (2018). - PubMed
-
- Sparber F., et al. ., The skin commensal yeast Malassezia triggers a type 17 response that coordinates anti-fungal immunity and exacerbates skin inflammation. Cell Host Microbe 25, 389–403.e6 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

