Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis
- PMID: 32576829
- PMCID: PMC7311409
- DOI: 10.1038/s41467-020-16983-2
Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis
Abstract
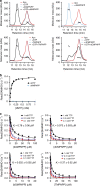
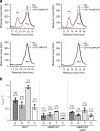
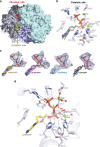
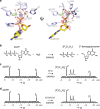
SAMHD1 regulates cellular 2'-deoxynucleoside-5'-triphosphate (dNTP) homeostasis by catalysing the hydrolysis of dNTPs into 2'-deoxynucleosides and triphosphate. In CD4+ myeloid lineage and resting T-cells, SAMHD1 blocks HIV-1 and other viral infections by depletion of the dNTP pool to a level that cannot support replication. SAMHD1 mutations are associated with the autoimmune disease Aicardi-Goutières syndrome and hypermutated cancers. Furthermore, SAMHD1 sensitises cancer cells to nucleoside-analogue anti-cancer therapies and is linked with DNA repair and suppression of the interferon response to cytosolic nucleic acids. Nevertheless, despite its requirement in these processes, the fundamental mechanism of SAMHD1-catalysed dNTP hydrolysis remained unknown. Here, we present structural and enzymological data showing that SAMHD1 utilises an active site, bi-metallic iron-magnesium centre that positions a hydroxide nucleophile in-line with the Pα-O5' bond to catalyse phosphoester bond hydrolysis. This precise molecular mechanism for SAMHD1 catalysis, reveals how SAMHD1 down-regulates cellular dNTP and modulates the efficacy of nucleoside-based anti-cancer and anti-viral therapies.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Li N, Zhang W, Cao X. Identification of human homologue of mouse IFN-gamma induced protein from human dendritic cells. Immunol. Lett. 2000;74:221–224. - PubMed
-
- Goldstone DC, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480:379–382. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

