Genome-wide DNA methylation analysis of KRAS mutant cell lines
- PMID: 32576853
- PMCID: PMC7311523
- DOI: 10.1038/s41598-020-66797-x
Genome-wide DNA methylation analysis of KRAS mutant cell lines
Abstract
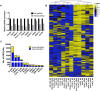
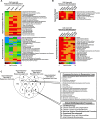
Oncogenic RAS mutations are associated with DNA methylation changes that alter gene expression to drive cancer. Recent studies suggest that DNA methylation changes may be stochastic in nature, while other groups propose distinct signaling pathways responsible for aberrant methylation. Better understanding of DNA methylation events associated with oncogenic KRAS expression could enhance therapeutic approaches. Here we analyzed the basal CpG methylation of 11 KRAS-mutant and dependent pancreatic cancer cell lines and observed strikingly similar methylation patterns. KRAS knockdown resulted in unique methylation changes with limited overlap between each cell line. In KRAS-mutant Pa16C pancreatic cancer cells, while KRAS knockdown resulted in over 8,000 differentially methylated (DM) CpGs, treatment with the ERK1/2-selective inhibitor SCH772984 showed less than 40 DM CpGs, suggesting that ERK is not a broadly active driver of KRAS-associated DNA methylation. KRAS G12V overexpression in an isogenic lung model reveals >50,600 DM CpGs compared to non-transformed controls. In lung and pancreatic cells, gene ontology analyses of DM promoters show an enrichment for genes involved in differentiation and development. Taken all together, KRAS-mediated DNA methylation are stochastic and independent of canonical downstream effector signaling. These epigenetically altered genes associated with KRAS expression could represent potential therapeutic targets in KRAS-driven cancer.
Conflict of interest statement
C.J.D. is on the Scientific Advisory Board of Mirati Therapeutics. C.J.D. received funding support from Mirati Therapeutics and Deciphera Pharmaceuticals. C.J.D. has been a consultant with Deciphera Pharmaceuticals, Eli Lilly, Jazz Therapeutics and Ribometrix.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

