Entropic Trapping of DNA with a Nanofiltered Nanopore
- PMID: 32577609
- PMCID: PMC7310961
- DOI: 10.1021/acsanm.9b00606
Entropic Trapping of DNA with a Nanofiltered Nanopore
Abstract
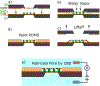
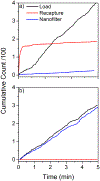
Elucidating the kinetics of DNA passage through a solid-state nanopore is a fertile field of research, and mechanisms for controlling capture, passage, and trapping of biopolymers are likely to find numerous technological applications. Here we present a nanofiltered nanopore device, which forms an entropic cage for DNA following first passage through the nanopore, trapping the translocated DNA and permitting recapture for subsequent reanalysis and investigation of kinetics of passage under confinement. We characterize the trapping properties of this nanodevice by driving individual DNA polymers into the nanoscale gap separating the nanofilter and the pore, forming an entropic cage similar to a "two pores in series" device, leaving polymers to diffuse in the cage for various time lengths, and attempting to recapture the same molecule. We show that the cage results in effectively permanent trapping when the radius of gyration of the target polymer is significantly larger than the radii of the pores in the nanofilter. We also compare translocation dynamics as a function of translocation direction in order to study the effects of confinement on DNA just prior to translocation, providing further insight into the nanopore translocation process. This nanofiltered nanopore device realizes simple fabrication of a femtoliter nanoreactor in which to study fundamental biophysics and biomolecular reactions on the single-molecule level. The device provides an electrically-permeable single-molecule trap with a higher entropic barrier to escape than previous attempts to fabricate similar structures.
Keywords: DNA; entropy; nanoconfinement; nanofabrication; nanopore; nanoporous membrane; nanotechnology.
Conflict of interest statement
Conflicts of Interest KB, GRM, JLM, and VTC declare competing financial interest in the form of a patent on the nanofiltered nanopore device. JLM is a cofounder of SiMPore Inc. All other authors declare no competing financial interest.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
