This is a preprint.
Structure-based Design of Prefusion-stabilized SARS-CoV-2 Spikes
- PMID: 32577660
- PMCID: PMC7302215
- DOI: 10.1101/2020.05.30.125484
Structure-based Design of Prefusion-stabilized SARS-CoV-2 Spikes
Update in
-
Structure-based design of prefusion-stabilized SARS-CoV-2 spikes.Science. 2020 Sep 18;369(6510):1501-1505. doi: 10.1126/science.abd0826. Epub 2020 Jul 23. Science. 2020. PMID: 32703906 Free PMC article.
Abstract
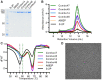
The COVID-19 pandemic caused by the novel coronavirus SARS-CoV-2 has led to accelerated efforts to develop therapeutics, diagnostics, and vaccines to mitigate this public health emergency. A key target of these efforts is the spike (S) protein, a large trimeric class I fusion protein that is metastable and difficult to produce recombinantly in large quantities. Here, we designed and expressed over 100 structure-guided spike variants based upon a previously determined cryo-EM structure of the prefusion SARS-CoV-2 spike. Biochemical, biophysical and structural characterization of these variants identified numerous individual substitutions that increased protein yields and stability. The best variant, HexaPro, has six beneficial proline substitutions leading to ~10-fold higher expression than its parental construct and is able to withstand heat stress, storage at room temperature, and multiple freeze-thaws. A 3.2 Å-resolution cryo-EM structure of HexaPro confirmed that it retains the prefusion spike conformation. High-yield production of a stabilized prefusion spike protein will accelerate the development of vaccines and serological diagnostics for SARS-CoV-2.
Conflict of interest statement
COMPETING INTERESTS N.W. and J.S.M. are inventors on U.S. patent application no. 62/412,703 (“Prefusion Coronavirus Spike Proteins and Their Use”). D.W., N.W. and J.S.M. are inventors on U.S. patent application no. 62/972,886 (“2019-nCoV Vaccine”). C.-L.H., J.A.G., J.M.S., C.-W.C., A.M.D., K.J., H.-C.K., D.W., P.O.B., C.K.H., N.V.J., N.W., J.A.M., I.J.F., and J.S.M. are inventors on U.S. patent application no. 63/032,502 (“Engineered Coronavirus Spike (S) Protein and Methods of Use Thereof”).
Figures

References
-
- Zaki A. M., Van Boheemen S., Bestebroer T. M., Osterhaus A. D. M. E. & Fouchier R. A. M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 (2012). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
