SFPQ and Tau: critical factors contributing to rapid progression of Alzheimer's disease
- PMID: 32577828
- PMCID: PMC7423812
- DOI: 10.1007/s00401-020-02178-y
SFPQ and Tau: critical factors contributing to rapid progression of Alzheimer's disease
Abstract
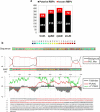
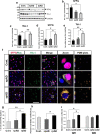
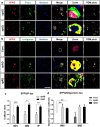
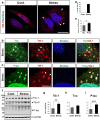
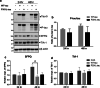
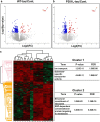
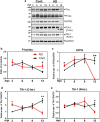
Dysfunctional RNA-binding proteins (RBPs) have been implicated in several neurodegenerative disorders. Recently, this paradigm of RBPs has been extended to pathophysiology of Alzheimer's disease (AD). Here, we identified disease subtype specific variations in the RNA-binding proteome (RBPome) of sporadic AD (spAD), rapidly progressive AD (rpAD), and sporadic Creutzfeldt Jakob disease (sCJD), as well as control cases using RNA pull-down assay in combination with proteomics. We show that one of these identified proteins, splicing factor proline and glutamine rich (SFPQ), is downregulated in the post-mortem brains of rapidly progressive AD patients, sCJD patients and 3xTg mice brain at terminal stage of the disease. In contrast, the expression of SFPQ was elevated at early stage of the disease in the 3xTg mice, and in vitro after oxidative stress stimuli. Strikingly, in rpAD patients' brains SFPQ showed a significant dislocation from the nucleus and cytoplasmic colocalization with TIA-1. Furthermore, in rpAD brain lesions, SFPQ and p-tau showed extranuclear colocalization. Of note, association between SFPQ and tau-oligomers in rpAD brains suggests a possible role of SFPQ in oligomerization and subsequent misfolding of tau protein. In line with the findings from the human brain, our in vitro study showed that SFPQ is recruited into TIA-1-positive stress granules (SGs) after oxidative stress induction, and colocalizes with tau/p-tau in these granules, providing a possible mechanism of SFPQ dislocation through pathological SGs. Furthermore, the expression of human tau in vitro induced significant downregulation of SFPQ, suggesting a causal role of tau in the downregulation of SFPQ. The findings from the current study indicate that the dysregulation and dislocation of SFPQ, the subsequent DNA-related anomalies and aberrant dynamics of SGs in association with pathological tau represents a critical pathway which contributes to rapid progression of AD.
Keywords: 3xTg mice; Dislocation; RNA-binding proteins; Rapidly progressive Alzheimer’s disease; SFPQ; Stress granules.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

