Screening of Chloroquine, Hydroxychloroquine and its derivatives for their binding affinity to multiple SARS-CoV-2 protein drug targets
- PMID: 32579059
- PMCID: PMC7332874
- DOI: 10.1080/07391102.2020.1782265
Screening of Chloroquine, Hydroxychloroquine and its derivatives for their binding affinity to multiple SARS-CoV-2 protein drug targets
Abstract
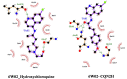
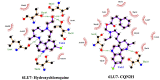
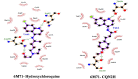
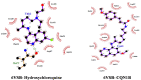
Recently Chloroquine and its derivative Hydroxychloroquine have garnered enormous interest amongst the clinicians and health authorities' world over as a potential treatment to contain COVID-19 pandemic. The present research aims at investigating the therapeutic potential of Chloroquine and its potent derivative Hydroxychloroquine against SARS-CoV-2 viral proteins. At the same time screening was performed for some chemically synthesized derivatives of Chloroquine and compared their binding efficacy with chemically synthesized Chloroquine derivatives through in silico approaches. For the purpose of the study, some essential viral proteins and enzymes were selected that are implicated in SARS-CoV-2 replication and multiplication as putative drug targets. Chloroquine, Hydroxychloroquine, and some of their chemically synthesized derivatives, taken from earlier published studies were selected as drug molecules. We have conducted molecular docking and related studies between Chloroquine and its derivatives and SARS-CoV-2 viral proteins, and the findings show that both Chloroquine and Hydroxychloroquine can bind to specific structural and non-structural proteins implicated in the pathogenesis of SARS-CoV-2 infection with different efficiencies. Our current study also shows that some of the chemically synthesized Chloroquine derivatives can also potentially inhibit various SARS-CoV-2 viral proteins by binding to them and concomitantly effectively disrupting the active site of these proteins. These findings bring into light another possible mechanism of action of Chloroquine and Hydroxychloroquine and also pave the way for further drug repurposing and remodeling.Communicated by Ramaswamy H. Sarma.
Keywords: COVID-19; Chloroquine derivatives; Hydroxychloroquine; SARS-CoV-2; molecular docking.
Figures


Similar articles
-
Docking study of chloroquine and hydroxychloroquine interaction with RNA binding domain of nucleocapsid phospho-protein - an in silico insight into the comparative efficacy of repurposing antiviral drugs.J Biomol Struct Dyn. 2021 Aug;39(12):4243-4255. doi: 10.1080/07391102.2020.1775703. Epub 2020 Jul 6. J Biomol Struct Dyn. 2021. PMID: 32469265
-
New Anti SARS-Cov-2 Targets for Quinoline Derivatives Chloroquine and Hydroxychloroquine.Int J Mol Sci. 2020 Aug 14;21(16):5856. doi: 10.3390/ijms21165856. Int J Mol Sci. 2020. PMID: 32824072 Free PMC article.
-
Approach to the mechanism of action of hydroxychloroquine on SARS-CoV-2: a molecular docking study.J Biomol Struct Dyn. 2021 Sep;39(15):5792-5798. doi: 10.1080/07391102.2020.1792993. Epub 2020 Jul 17. J Biomol Struct Dyn. 2021. PMID: 32677545 Free PMC article.
-
Insights into antiviral mechanisms of remdesivir, lopinavir/ritonavir and chloroquine/hydroxychloroquine affecting the new SARS-CoV-2.Biomed Pharmacother. 2020 Nov;131:110668. doi: 10.1016/j.biopha.2020.110668. Epub 2020 Aug 24. Biomed Pharmacother. 2020. PMID: 32861965 Free PMC article. Review.
-
A review on possible modes of action of chloroquine/hydroxychloroquine: repurposing against SAR-CoV-2 (COVID-19) pandemic.Int J Antimicrob Agents. 2020 Aug;56(2):106028. doi: 10.1016/j.ijantimicag.2020.106028. Epub 2020 May 22. Int J Antimicrob Agents. 2020. PMID: 32450198 Free PMC article. Review.
Cited by
-
Diminazene Aceturate Reduces Angiotensin II Constriction and Interacts with the Spike Protein of Severe Acute Respiratory Syndrome Coronavirus 2.Biomedicines. 2022 Jul 18;10(7):1731. doi: 10.3390/biomedicines10071731. Biomedicines. 2022. PMID: 35885036 Free PMC article.
-
Atorvastatin Effectively Inhibits Ancestral and Two Emerging Variants of SARS-CoV-2 in vitro.Front Microbiol. 2022 Mar 18;13:721103. doi: 10.3389/fmicb.2022.721103. eCollection 2022. Front Microbiol. 2022. PMID: 35369500 Free PMC article.
-
Repurposing FDA-approved drugs against multiple proteins of SARS-CoV-2: An in silico study.Sci Afr. 2021 Sep;13:e00845. doi: 10.1016/j.sciaf.2021.e00845. Epub 2021 Jul 11. Sci Afr. 2021. PMID: 34308004 Free PMC article.
-
Understanding the Driving Forces That Trigger Mutations in SARS-CoV-2: Mutational Energetics and the Role of Arginine Blockers in COVID-19 Therapy.Viruses. 2022 May 11;14(5):1029. doi: 10.3390/v14051029. Viruses. 2022. PMID: 35632769 Free PMC article.
-
BPR3P0128, a non-nucleoside RNA-dependent RNA polymerase inhibitor, inhibits SARS-CoV-2 variants of concern and exerts synergistic antiviral activity in combination with remdesivir.Antimicrob Agents Chemother. 2024 Apr 3;68(4):e0095623. doi: 10.1128/aac.00956-23. Epub 2024 Mar 6. Antimicrob Agents Chemother. 2024. PMID: 38446062 Free PMC article.
References
-
- Bommu U. D., Konidala K. K., Pamanji R., & Yeguvapalli S. (2019). Structural probing, screening and structure-based drug repositioning insights into the identification of potential Cox-2 inhibitors from selective coxibs. Interdisciplinary Sciences: Computational Life Sciences, 11(2), 153–169. 10.1007/s12539-017-0244-5 - DOI - PubMed
-
- CEBM (2020). Global Covid-19 case fatality rates Retrieved May 7, 2020, from https://www.cebm.net/covid-19/global-covid-19-case-fatality-rates/.
-
- DeLano W. L. (2002). Pymol: An open-source molecular graphics tool. CCP4 Newsletter on Protein Crystallography, 40(1), 82–92.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
