Pharmacoinformatics and molecular dynamics simulation studies reveal potential covalent and FDA-approved inhibitors of SARS-CoV-2 main protease 3CLpro
- PMID: 32579061
- PMCID: PMC7332866
- DOI: 10.1080/07391102.2020.1782768
Pharmacoinformatics and molecular dynamics simulation studies reveal potential covalent and FDA-approved inhibitors of SARS-CoV-2 main protease 3CLpro
Abstract
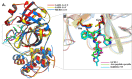
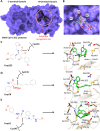
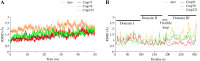
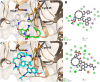
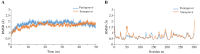
The SARS-CoV-2 was confirmed to cause the global pandemic of coronavirus disease 2019 (COVID-19). The 3-chymotrypsin-like protease (3CLpro), an essential enzyme for viral replication, is a valid target to combat SARS-CoV and MERS-CoV. In this work, we present a structure-based study to identify potential covalent inhibitors containing a variety of chemical warheads. The targeted Asinex Focused Covalent (AFCL) library was screened based on different reaction types and potential covalent inhibitors were identified. In addition, we screened FDA-approved protease inhibitors to find candidates to be repurposed against SARS-CoV-2 3CLpro. A number of compounds with significant covalent docking scores were identified. These compounds were able to establish a covalent bond (C-S) with the reactive thiol group of Cys145 and to form favorable interactions with residues lining the substrate-binding site. Moreover, paritaprevir and simeprevir from FDA-approved protease inhibitors were identified as potential inhibitors of SARS-CoV-2 3CLpro. The mechanism and dynamic stability of binding between the identified compounds and SARS-CoV-2 3CLpro were characterized by molecular dynamics (MD) simulations. The identified compounds are potential inhibitors worthy of further development as COVID-19 drugs. Importantly, the identified FDA-approved anti-hepatitis-C virus (HCV) drugs paritaprevir and simeprevir could be ready for clinical trials to treat infected patients and help curb COVID-19. Communicated by Ramaswamy H. Sarma.
Keywords: 3CL protease; COVID-19; SARS-CoV-2; covalent inhibitors; molecular dynamics simulation; paritaprevir; simeprevir.
Figures

References
-
- Ahmed B., Ali Ashfaq U., & Usman Mirza M. (2018). Medicinal plant phytochemicals and their inhibitory activities against pancreatic lipase: Molecular docking combined with molecular dynamics simulation approach. Natural Product Research, 32(10), 1123–1129. 10.1080/14786419.2017.1320786 - DOI - PubMed
-
- Ahmed T., Noman M., Almatroudi A., Shahid M., Khurshid M., Tariq F., Ul Qamar M. T., Yu R., & Li B. (2020). Coronavirus disease 2019 assosiated pneumonia in China: Current status and future prospects. Preprints, 2020020358. 10.20944/preprints202002.0358.v3 - DOI
-
- Berman H. M., Bourne P. E., Westbrook J., & Zardecki C. (2003). The protein data bank In Protein structure (pp. 394–410). CRC Press.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
