Platon: identification and characterization of bacterial plasmid contigs in short-read draft assemblies exploiting protein sequence-based replicon distribution scores
- PMID: 32579097
- PMCID: PMC7660248
- DOI: 10.1099/mgen.0.000398
Platon: identification and characterization of bacterial plasmid contigs in short-read draft assemblies exploiting protein sequence-based replicon distribution scores
Abstract
Plasmids are extrachromosomal genetic elements that replicate independently of the chromosome and play a vital role in the environmental adaptation of bacteria. Due to potential mobilization or conjugation capabilities, plasmids are important genetic vehicles for antimicrobial resistance genes and virulence factors with huge and increasing clinical implications. They are therefore subject to large genomic studies within the scientific community worldwide. As a result of rapidly improving next-generation sequencing methods, the quantity of sequenced bacterial genomes is constantly increasing, in turn raising the need for specialized tools to (i) extract plasmid sequences from draft assemblies, (ii) derive their origin and distribution, and (iii) further investigate their genetic repertoire. Recently, several bioinformatic methods and tools have emerged to tackle this issue; however, a combination of high sensitivity and specificity in plasmid sequence identification is rarely achieved in a taxon-independent manner. In addition, many software tools are not appropriate for large high-throughput analyses or cannot be included in existing software pipelines due to their technical design or software implementation. In this study, we investigated differences in the replicon distributions of protein-coding genes on a large scale as a new approach to distinguish plasmid-borne from chromosome-borne contigs. We defined and computed statistical discrimination thresholds for a new metric: the replicon distribution score (RDS), which achieved an accuracy of 96.6 %. The final performance was further improved by the combination of the RDS metric with heuristics exploiting several plasmid-specific higher-level contig characterizations. We implemented this workflow in a new high-throughput taxon-independent bioinformatics software tool called Platon for the recruitment and characterization of plasmid-borne contigs from short-read draft assemblies. Compared to PlasFlow, Platon achieved a higher accuracy (97.5 %) and more balanced predictions (F1=82.6 %) tested on a broad range of bacterial taxa and better or equal performance against the targeted tools PlasmidFinder and PlaScope on sequenced Escherichia coli isolates. Platon is available at: http://platon.computational.bio/.
Keywords: whole-genome sequencing; NGS; bacteria; plasmids.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



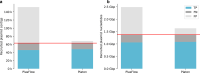

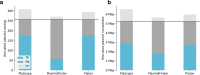
References
-
- Clark DP, Stahl DA, Martinko JM, Madigan MT. Brock biology of microorganisms (13th edition). Benjamin Cummings. 2010 https://www.amazon.com/Brock-Biology-Microorganisms-Michael-Madigan/dp/0...
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

