SARS-CoV-2 and SARS-CoV: Virtual screening of potential inhibitors targeting RNA-dependent RNA polymerase activity (NSP12)
- PMID: 32579254
- PMCID: PMC7361265
- DOI: 10.1002/jmv.26222
SARS-CoV-2 and SARS-CoV: Virtual screening of potential inhibitors targeting RNA-dependent RNA polymerase activity (NSP12)
Abstract
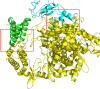
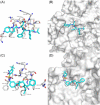
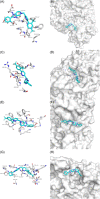
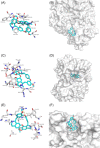
Since the outbreak of severe acute respiratory syndrome (SARS) in 2003, the harm caused by coronaviruses to the world cannot be underestimated. Recently, a novel coronavirus (severe acute respiratory syndrome coronavirus-2 [SARS-CoV-2]) initially found to trigger human severe respiratory illness in Wuhan City of China in 2019, has infected more than six million people worldwide by 21 June 2020, and which has been recognized as a public health emergency of international concern as well. And the virus has spread to more than 200 countries around the world. However, the effective drug has not yet been officially licensed or approved to treat SARS-Cov-2 and SARS-Cov infection. NSP12-NSP7-NSP8 complex of SARS-CoV-2 or SARS-CoV, essential for viral replication and transcription, is generally regarded as a potential target to fight against the virus. According to the NSP12-NSP7-NSP8 complex (PDB ID: 7BW4) structure of SARS-CoV-2 and the NSP12-NSP7-NSP8 complex (PDB ID: 6NUR) structure of SARS-CoV, NSP12-NSP7 interface model, and NSP12-NSP8 interface model were established for virtual screening in the present study. Eight compounds (Nilotinib, Saquinavir, Tipranavir, Lonafarnib, Tegobuvir, Olysio, Filibuvir, and Cepharanthine) were selected for binding free energy calculations based on virtual screening and docking scores. All eight compounds can combine well with NSP12-NSP7-NSP8 in the crystal structure, providing drug candidates for the treatment and prevention of coronavirus disease 2019 and SARS.
Keywords: COVID-19; NSP12-NSP7-NSP8; SARS-CoV; SARS-CoV-2; antiviral drugs; drug candidates.
© 2020 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare that there are no conflict of interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

