Nuclear body phase separation drives telomere clustering in ALT cancer cells
- PMID: 32579423
- PMCID: PMC7543070
- DOI: 10.1091/mbc.E19-10-0589
Nuclear body phase separation drives telomere clustering in ALT cancer cells
Abstract
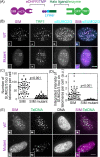
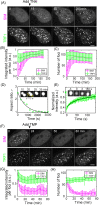
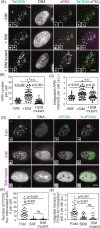
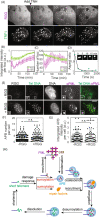
Telomerase-free cancer cells employ a recombination-based alternative lengthening of telomeres (ALT) pathway that depends on ALT-associated promyelocytic leukemia nuclear bodies (APBs), whose function is unclear. We find that APBs behave as liquid condensates in response to telomere DNA damage, suggesting two potential functions: condensation to enrich DNA repair factors and coalescence to cluster telomeres. To test these models, we developed a chemically induced dimerization approach to induce de novo APB condensation in live cells without DNA damage. We show that telomere-binding protein sumoylation nucleates APB condensation via interactions between small ubiquitin-like modifier (SUMO) and SUMO interaction motif (SIM), and that APB coalescence drives telomere clustering. The induced APBs lack DNA repair factors, indicating that APB functions in promoting telomere clustering can be uncoupled from enriching DNA repair factors. Indeed, telomere clustering relies only on liquid properties of the condensate, as an alternative condensation chemistry also induces clustering independent of sumoylation. Our findings introduce a chemical dimerization approach to manipulate phase separation and demonstrate how the material properties and chemical composition of APBs independently contribute to ALT, suggesting a general framework for how chromatin condensates promote cellular functions.
Figures

Similar articles
-
SUMO promotes DNA repair protein collaboration to support alternative telomere lengthening in the absence of PML.Genes Dev. 2024 Aug 20;38(13-14):614-630. doi: 10.1101/gad.351667.124. Genes Dev. 2024. PMID: 39038850 Free PMC article.
-
PML induces compaction, TRF2 depletion and DNA damage signaling at telomeres and promotes their alternative lengthening.J Cell Sci. 2015 May 15;128(10):1887-900. doi: 10.1242/jcs.148296. Epub 2015 Apr 23. J Cell Sci. 2015. PMID: 25908860
-
De novo assembly of a PML nuclear subcompartment occurs through multiple pathways and induces telomere elongation.J Cell Sci. 2011 Nov 1;124(Pt 21):3603-18. doi: 10.1242/jcs.084681. Epub 2011 Nov 1. J Cell Sci. 2011. PMID: 22045732
-
Biomolecular Condensates in Telomere Maintenance of ALT Cancer Cells.J Mol Biol. 2025 Mar 1;437(5):168951. doi: 10.1016/j.jmb.2025.168951. Epub 2025 Jan 16. J Mol Biol. 2025. PMID: 39826712 Review.
-
PML body meets telomere: the beginning of an ALTernate ending?Nucleus. 2012 May-Jun;3(3):263-75. doi: 10.4161/nucl.20326. Epub 2012 May 1. Nucleus. 2012. PMID: 22572954 Free PMC article. Review.
Cited by
-
Double-strand break repair and mis-repair in 3D.DNA Repair (Amst). 2023 Jan;121:103430. doi: 10.1016/j.dnarep.2022.103430. Epub 2022 Nov 17. DNA Repair (Amst). 2023. PMID: 36436496 Free PMC article.
-
Phase separations in oncogenesis, tumor progressions and metastasis: a glance from hallmarks of cancer.J Hematol Oncol. 2023 Dec 18;16(1):123. doi: 10.1186/s13045-023-01522-5. J Hematol Oncol. 2023. PMID: 38110976 Free PMC article. Review.
-
Melatonin: Regulation of Prion Protein Phase Separation in Cancer Multidrug Resistance.Molecules. 2022 Jan 21;27(3):705. doi: 10.3390/molecules27030705. Molecules. 2022. PMID: 35163973 Free PMC article. Review.
-
Phase Separation in Biology and Disease; Current Perspectives and Open Questions.J Mol Biol. 2023 Mar 1;435(5):167971. doi: 10.1016/j.jmb.2023.167971. Epub 2023 Jan 21. J Mol Biol. 2023. PMID: 36690068 Free PMC article.
-
Alternative Lengthening of Telomeres and Mediated Telomere Synthesis.Cancers (Basel). 2022 Apr 27;14(9):2194. doi: 10.3390/cancers14092194. Cancers (Basel). 2022. PMID: 35565323 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

