Identification of an immune-related long non-coding RNA signature and nomogram as prognostic target for muscle-invasive bladder cancer
- PMID: 32579540
- PMCID: PMC7343518
- DOI: 10.18632/aging.103369
Identification of an immune-related long non-coding RNA signature and nomogram as prognostic target for muscle-invasive bladder cancer
Abstract
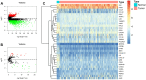
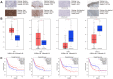
To identify an immune-related prognostic signature based on long non-coding RNAs (lncRNAs) and find immunotherapeutic targets for bladder urothelial carcinoma, we downloaded RNA-sequencing data from The Cancer Genome Atlas (TCGA) dataset. Functional enrichment analysis demonstrated bladder urothelial carcinoma was related to immune-related functions. We obtained 332 immune-related genes and 262 lncRNAs targeting immune-related genes. We constructed a signature based on eight lncRNAs in training cohort. Patients were classified as high-risk and low-risk according to signature risk score. High-risk patients had poor overall survival compared with low-risk patients (P < 0.001). Multivariate Cox regression suggested the signature was an independent prognostic indicator. The findings were further validated in testing, entire TCGA and external validation cohorts. Gene set enrichment analysis indicated significant enrichment of immune-related phenotype in high-risk group. Immunohistochemistry and online analyses validated the functions of 4 key immune-related genes (LIG1, TBX1, CTSG and CXCL12) in bladder urothelial carcinoma. Nomogram proved to be a good classifier for muscle-invasive bladder cancer through combining the signature. In conclusion, our immune-related prognostic signature and nomogram provided prognostic indicators and potential immunotherapeutic targets for muscle-invasive bladder cancer.
Keywords: immune-related; lncRNA signature; muscle-invasive bladder cancer; nomogram; prognostic model.
Conflict of interest statement
Figures









References
-
- Ebrahimi H, Amini E, Pishgar F, Moghaddam SS, Nabavizadeh B, Rostamabadi Y, Aminorroaya A, Fitzmaurice C, Farzadfar F, Nowroozi MR, Black PC, Daneshmand S. Global, regional and national burden of bladder cancer, 1990 to 2016: results from the GBD study 2016. J Urol. 2019; 201:893–901. 10.1097/JU.0000000000000025 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

