Signature genes associated with immunological non-responsiveness to anti-retroviral therapy in HIV-1 subtype-c infection
- PMID: 32579550
- PMCID: PMC7313746
- DOI: 10.1371/journal.pone.0234270
Signature genes associated with immunological non-responsiveness to anti-retroviral therapy in HIV-1 subtype-c infection
Abstract
Objective: HIV-infected individuals undergoing therapy may show an immunological-discordant response to therapy, with poor CD4+ T cells recovery, despite viral suppression below the detection limit. The present study was carried out to delineate the underlying molecular mechanisms of immunological non-responsiveness to HIV therapy.
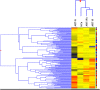
Design: We conducted microarray-based whole gene expression profiles of 30 subjects infected with HIV-1 subtype C, in peripheral blood to discern the signature genes associated with immunological non-responsiveness. After a thorough analysis and comparison of gene-expression profiles, microarray data was validated via qRT-PCR approach.
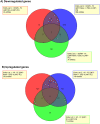
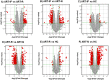
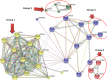
Results: Overall, we found 10 genes significantly up-regulated and 60 genes down-regulated (≥2-fold change) in immunological non-responders as compared to responders. Based on these results and pathway analysis of the protein-protein interaction, 20 genes were shortlisted for validation in human infected cases. We found statistically significant differences in expression levels of twelve genes IL-1α, IL-1β, IL-7R, TNF-α, FoxP3, PDCD5, COX7B, SOCS1, SOCS3, RPL9, RPL23, and LRRN3 respectively among immunological non-responders compared to therapy responders, confirming their an intimate relationship with immunological responsiveness to therapy.
Conclusions: Altogether, microarray and qRT-PCR validation results indicated that the aberrant expression of key genes involved in the regulation of T cell homeostasis, immune activation, inflammatory cytokine production, apoptosis, and immune-regulatory processes are possibly associated with immunological non-responsiveness in HIV-1 C infected individuals on ART.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

