A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids
- PMID: 32579880
- PMCID: PMC7303620
- DOI: 10.1016/j.stem.2020.06.015
A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids
Abstract
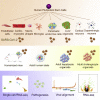
SARS-CoV-2 has caused the COVID-19 pandemic. There is an urgent need for physiological models to study SARS-CoV-2 infection using human disease-relevant cells. COVID-19 pathophysiology includes respiratory failure but involves other organ systems including gut, liver, heart, and pancreas. We present an experimental platform comprised of cell and organoid derivatives from human pluripotent stem cells (hPSCs). A Spike-enabled pseudo-entry virus infects pancreatic endocrine cells, liver organoids, cardiomyocytes, and dopaminergic neurons. Recent clinical studies show a strong association with COVID-19 and diabetes. We find that human pancreatic beta cells and liver organoids are highly permissive to SARS-CoV-2 infection, further validated using adult primary human islets and adult hepatocyte and cholangiocyte organoids. SARS-CoV-2 infection caused striking expression of chemokines, as also seen in primary human COVID-19 pulmonary autopsy samples. hPSC-derived cells/organoids provide valuable models for understanding the cellular responses of human tissues to SARS-CoV-2 infection and for disease modeling of COVID-19.
Keywords: SARS-CoV-2; alpha cells; beta cells; human pluripotent stem cells; liver organoids; pancreatic endocrine cells.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests R.E.S. is on the scientific advisory board of Miromatrix Inc. The other authors have no conflicts of interest.
Figures


Comment in
-
Mounting clues suggest the coronavirus might trigger diabetes.Nature. 2020 Jul;583(7814):16-17. doi: 10.1038/d41586-020-01891-8. Nature. 2020. PMID: 32606460 No abstract available.
-
Commentary: A Human Pluripotent Stem Cell-Based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids.Front Endocrinol (Lausanne). 2020 Oct 14;11:585922. doi: 10.3389/fendo.2020.585922. eCollection 2020. Front Endocrinol (Lausanne). 2020. PMID: 33162939 Free PMC article. No abstract available.
References
-
- Anagandula M., Richardson S.J., Oberste M.S., Sioofy-Khojine A.B., Hyöty H., Morgan N.G., Korsgren O., Frisk G. Infection of human islets of Langerhans with two strains of Coxsackie B virus serotype 1: assessment of virus replication, degree of cell death and induction of genes involved in the innate immunity pathway. J. Med. Virol. 2014;86:1402–1411. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R35 HL135778/HL/NHLBI NIH HHS/United States
- R01 AG056298/AG/NIA NIH HHS/United States
- R00 CA226353/CA/NCI NIH HHS/United States
- R01 CA234614/CA/NCI NIH HHS/United States
- F30 MH115616/MH/NIMH NIH HHS/United States
- R01 DK124463/DK/NIDDK NIH HHS/United States
- R03 DK117252/DK/NIDDK NIH HHS/United States
- UC4 DK112217/DK/NIDDK NIH HHS/United States
- R01 DK121072/DK/NIDDK NIH HHS/United States
- R01 AI107301/AI/NIAID NIH HHS/United States
- T32 AI007647/AI/NIAID NIH HHS/United States
- DP3 DK111907/DK/NIDDK NIH HHS/United States
- R01 DK119667/DK/NIDDK NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R01 DK116075/DK/NIDDK NIH HHS/United States
- K99 CA226353/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

