mTOR Signaling and SREBP Activity Increase FADS2 Expression and Can Activate Sapienate Biosynthesis
- PMID: 32579932
- PMCID: PMC7326293
- DOI: 10.1016/j.celrep.2020.107806
mTOR Signaling and SREBP Activity Increase FADS2 Expression and Can Activate Sapienate Biosynthesis
Abstract
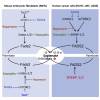
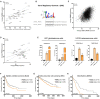
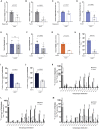
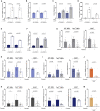
Cancer cells display an increased plasticity in their lipid metabolism, which includes the conversion of palmitate to sapienate via the enzyme fatty acid desaturase 2 (FADS2). We find that FADS2 expression correlates with mammalian target of rapamycin (mTOR) signaling and sterol regulatory element-binding protein 1 (SREBP-1) activity across multiple cancer types and is prognostic in some cancer types. Accordingly, activating mTOR signaling by deleting tuberous sclerosis complex 2 (Tsc2) or overexpression of SREBP-1/2 is sufficient to increase FADS2 mRNA expression and sapienate metabolism in mouse embryonic fibroblasts (MEFs) and U87 glioblastoma cells, respectively. Conversely, inhibiting mTOR signaling decreases FADS2 expression and sapienate biosynthesis in MEFs with Tsc2 deletion, HUH7 hepatocellular carcinoma cells, and orthotopic HUH7 liver xenografts. In conclusion, we show that mTOR signaling and SREBP activity are sufficient to activate sapienate metabolism by increasing FADS2 expression. Consequently, targeting mTOR signaling can reduce sapienate metabolism in vivo.
Keywords: FADS2; SCD1; SREBP; cancer; fatty acid metabolism; glioblastoma; hepatocellular carcinoma; mTOR; palmitate; palmitoleate; sapienate.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests S.-M.F. has received funding from Bayer, Merck, and Black Belt Therapeutics and has consulted for Funds+.
Figures


References
-
- Cho H.P., Nakamura M.T., Clarke S.D. Cloning, expression, and nutritional regulation of the mammalian Δ-6 desaturase. J. Biol. Chem. 1999;274:471–477. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

