Learning Representations to Predict Intermolecular Interactions on Large-Scale Heterogeneous Molecular Association Network
- PMID: 32580123
- PMCID: PMC7317230
- DOI: 10.1016/j.isci.2020.101261
Learning Representations to Predict Intermolecular Interactions on Large-Scale Heterogeneous Molecular Association Network
Abstract
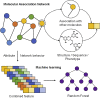
Molecular components that are functionally interdependent in human cells constitute molecular association networks. Disease can be caused by disturbance of multiple molecular interactions. New biomolecular regulatory mechanisms can be revealed by discovering new biomolecular interactions. To this end, a heterogeneous molecular association network is formed by systematically integrating comprehensive associations between miRNAs, lncRNAs, circRNAs, mRNAs, proteins, drugs, microbes, and complex diseases. We propose a machine learning method for predicting intermolecular interactions, named MMI-Pred. More specifically, a network embedding model is developed to fully exploit the network behavior of biomolecules, and attribute features are also calculated. Then, these discriminative features are combined to train a random forest classifier to predict intermolecular interactions. MMI-Pred achieves an outstanding performance of 93.50% accuracy in hybrid associations prediction under 5-fold cross-validation. This work provides systematic landscape and machine learning method to model and infer complex associations between various biological components.
Keywords: Biocomputational Method; Bioinformatics; Computational Bioinformatics.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures





Similar articles
-
Construction and Analysis of Molecular Association Network by Combining Behavior Representation and Node Attributes.Front Genet. 2019 Nov 7;10:1106. doi: 10.3389/fgene.2019.01106. eCollection 2019. Front Genet. 2019. PMID: 31788002 Free PMC article.
-
Learning Representation of Molecules in Association Network for Predicting Intermolecular Associations.IEEE/ACM Trans Comput Biol Bioinform. 2021 Nov-Dec;18(6):2546-2554. doi: 10.1109/TCBB.2020.2973091. Epub 2021 Dec 8. IEEE/ACM Trans Comput Biol Bioinform. 2021. PMID: 32070992
-
NEMPD: a network embedding-based method for predicting miRNA-disease associations by preserving behavior and attribute information.BMC Bioinformatics. 2020 Sep 10;21(1):401. doi: 10.1186/s12859-020-03716-x. BMC Bioinformatics. 2020. PMID: 32912137 Free PMC article.
-
Integrative Construction and Analysis of Molecular Association Network in Human Cells by Fusing Node Attribute and Behavior Information.Mol Ther Nucleic Acids. 2020 Mar 6;19:498-506. doi: 10.1016/j.omtn.2019.10.046. Epub 2019 Nov 29. Mol Ther Nucleic Acids. 2020. PMID: 31923739 Free PMC article.
-
Machine learning approaches for predicting biomolecule-disease associations.Brief Funct Genomics. 2021 Jul 17;20(4):273-287. doi: 10.1093/bfgp/elab002. Brief Funct Genomics. 2021. PMID: 33554238 Review.
Cited by
-
iPiDA-SWGCN: Identification of piRNA-disease associations based on Supplementarily Weighted Graph Convolutional Network.PLoS Comput Biol. 2023 Jun 20;19(6):e1011242. doi: 10.1371/journal.pcbi.1011242. eCollection 2023 Jun. PLoS Comput Biol. 2023. PMID: 37339125 Free PMC article.
-
A potential therapeutic molecule target: lncRNA AK023507 inhibits the metastasis of breast cancer by regulating the WNT/DOCK4/β-catenin axis.Breast Cancer Res Treat. 2025 Jun;211(3):727-741. doi: 10.1007/s10549-025-07695-6. Epub 2025 Apr 9. Breast Cancer Res Treat. 2025. PMID: 40205246
-
DANE-MDA: Predicting microRNA-disease associations via deep attributed network embedding.iScience. 2021 Apr 20;24(6):102455. doi: 10.1016/j.isci.2021.102455. eCollection 2021 Jun 25. iScience. 2021. PMID: 34041455 Free PMC article.
-
An effective drug-disease associations prediction model based on graphic representation learning over multi-biomolecular network.BMC Bioinformatics. 2022 Jan 4;23(1):9. doi: 10.1186/s12859-021-04553-2. BMC Bioinformatics. 2022. PMID: 34983364 Free PMC article.
-
Prediction of miRNA-disease associations based on PCA and cascade forest.BMC Bioinformatics. 2024 Dec 19;25(1):386. doi: 10.1186/s12859-024-05999-w. BMC Bioinformatics. 2024. PMID: 39701957 Free PMC article.
References
-
- Ay M., Goh K.-I., Cusick M.E., Barabasi A.-L., Vidal M. Drug--target network. Nat. Biotechnol. 2007;25:1119–1127. - PubMed
-
- Bandres E., Agirre X., Bitarte N., Ramirez N., Zarate R., Romangomez J., Prosper F., Garciafoncillas J. Epigenetic regulation of microRNA expression in colorectal cancer. Int. J. Cancer. 2009;125:2737–2743. - PubMed
-
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
LinkOut - more resources
Full Text Sources

