Discovery of CD80 and CD86 as recent activation markers on regulatory T cells by protein-RNA single-cell analysis
- PMID: 32580776
- PMCID: PMC7315544
- DOI: 10.1186/s13073-020-00756-z
Discovery of CD80 and CD86 as recent activation markers on regulatory T cells by protein-RNA single-cell analysis
Abstract
Background: Traditionally, the transcriptomic and proteomic characterisation of CD4+ T cells at the single-cell level has been performed by two largely exclusive types of technologies: single-cell RNA sequencing (scRNA-seq) and antibody-based cytometry. Here, we present a multi-omics approach allowing the simultaneous targeted quantification of mRNA and protein expression in single cells and investigate its performance to dissect the heterogeneity of human immune cell populations.
Methods: We have quantified the single-cell expression of 397 genes at the mRNA level and up to 68 proteins using oligo-conjugated antibodies (AbSeq) in 43,656 primary CD4+ T cells isolated from the blood and 31,907 CD45+ cells isolated from the blood and matched duodenal biopsies. We explored the sensitivity of this targeted scRNA-seq approach to dissect the heterogeneity of human immune cell populations and identify trajectories of functional T cell differentiation.
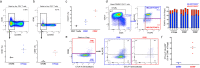
Results: We provide a high-resolution map of human primary CD4+ T cells and identify precise trajectories of Th1, Th17 and regulatory T cell (Treg) differentiation in the blood and tissue. The sensitivity provided by this multi-omics approach identified the expression of the B7 molecules CD80 and CD86 on the surface of CD4+ Tregs, and we further demonstrated that B7 expression has the potential to identify recently activated T cells in circulation. Moreover, we identified a rare subset of CCR9+ T cells in the blood with tissue-homing properties and expression of several immune checkpoint molecules, suggestive of a regulatory function.
Conclusions: The transcriptomic and proteomic hybrid technology described in this study provides a cost-effective solution to dissect the heterogeneity of immune cell populations at extremely high resolution. Unexpectedly, CD80 and CD86, normally expressed on antigen-presenting cells, were detected on a subset of activated Tregs, indicating a role for these co-stimulatory molecules in regulating the dynamics of CD4+ T cell responses.
Keywords: AbSeq; C-C chemokine receptor type 9 (CCR9); CD4+ T cells; CD80; CD86; Immunophenotyping; Multi-omics; Regulatory T cells (Tregs); Single-cell RNA sequencing (scRNA-seq).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







References
-
- Ornatsky O, Bandura D, Baranov V, Nitz M, Winnik MA, Tanner S. Highly multiparametric analysis by mass cytometry. J Immunol Methods. 2010;361:1–20. - PubMed
-
- Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2017;18:35. - PubMed
-
- Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

