A single cell transcriptional atlas of early synovial joint development
- PMID: 32580935
- PMCID: PMC7390639
- DOI: 10.1242/dev.185777
A single cell transcriptional atlas of early synovial joint development
Abstract
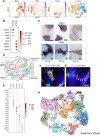
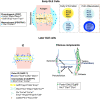
Synovial joint development begins with the formation of the interzone, a region of condensed mesenchymal cells at the site of the prospective joint. Recently, lineage-tracing strategies have revealed that Gdf5-lineage cells native to and from outside the interzone contribute to most, if not all, of the major joint components. However, there is limited knowledge of the specific transcriptional and signaling programs that regulate interzone formation and fate diversification of synovial joint constituents. To address this, we have performed single cell RNA-Seq analysis of 7329 synovial joint progenitor cells from the developing murine knee joint from E12.5 to E15.5. By using a combination of computational analytics, in situ hybridization and in vitro characterization of prospectively isolated populations, we have identified the transcriptional profiles of the major developmental paths for joint progenitors. Our freely available single cell transcriptional atlas will serve as a resource for the community to uncover transcriptional programs and cell interactions that regulate synovial joint development.
Keywords: Articular cartilage; Chondrocyte; Gdf5; Interzone; Ligament; Meniscus; Single cell RNA-Seq; Synovial joint development; Synovium.
© 2020. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures





References
-
- Aro E., Salo A. M., Khatri R., Finnilä M., Miinalainen I., Sormunen R., Pakkanen O., Holster T., Soininen R., Prein C. et al. (2015). Severe extracellular matrix abnormalities and chondrodysplasia in mice lacking collagen Prolyl 4-hydroxylase isoenzyme II in combination with a reduced amount of Isoenzyme I. J. Biol. Chem. 290, 16964-16978. 10.1074/jbc.M115.662635 - DOI - PMC - PubMed
-
- Blackburn P. R., Xu Z., Tumelty K. E., Zhao R. W., Monis W. J., Harris K. G., Gass J. M., Cousin M. A., Boczek N. J., Mitkov M. V. et al. (2018). Bi-allelic alterations in AEBP1 lead to defective collagen assembly and connective tissue structure resulting in a variant of ehlers-danlos syndrome. Am. J. Hum. Genet. 102, 696-705. 10.1016/j.ajhg.2018.02.018 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

