Induction of Rod-Shaped Structures by Herpes Simplex Virus Glycoprotein I
- PMID: 32581097
- PMCID: PMC7431791
- DOI: 10.1128/JVI.00231-20
Induction of Rod-Shaped Structures by Herpes Simplex Virus Glycoprotein I
Abstract
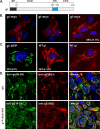
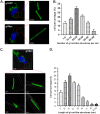
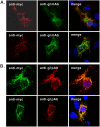
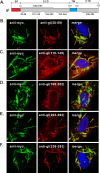
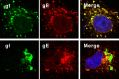
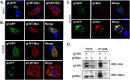
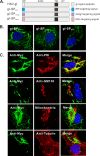
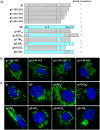
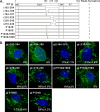
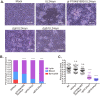
The envelope glycoprotein I (gI) of herpes simplex virus 1 (HSV-1) is a critical mediator of virus-induced cell-to-cell spread and cell-cell fusion. Here, we report a previously unrecognized property of this molecule. In transfected cells, the HSV-1 gI was discovered to induce rod-shaped structures that were uniform in width but variable in length. Moreover, the gI within these structures was conformationally different from the typical form of gI, as a previously used monoclonal antibody mAb3104 and a newly made peptide antibody to the gI extracellular domain (ECD) (amino acids [aa] 110 to 202) both failed to stain the long rod-shaped structures, suggesting the formation of a higher-order form. Consistent with this observation, we found that gI could self-interact and that the rod-shaped structures failed to recognize glycoprotein E, the well-known binding partner of gI. Further analyses by deletion mutagenesis and construction of chimeric mutants between gI and gD revealed that the gI ECD is the critical determinant, whereas the transmembrane domain served merely as an anchor. The critical amino acids were subsequently mapped to proline residues 184 and 188 within a conserved PXXXP motif. Reverse genetics analyses showed that the ability to induce a rod-shaped structure was not required for viral replication and spread in cell culture but rather correlated positively with the capability of the virus to induce cell fusion in the UL24syn background. Together, this work discovered a novel feature of HSV-1 gI that may have important implications in understanding gI function in viral spread and pathogenesis.IMPORTANCE The HSV-1 gI is required for viral cell-to-cell spread within the host, but the molecular mechanisms of how gI exactly works have remained poorly understood. Here, we report a novel property of this molecule, namely, induction of rod-shaped structures, which appeared to represent a higher-order form of gI. We further mapped the critical residues and showed that the ability of gI to induce rod-shaped structures correlated well with the capability of HSV-1 to induce cell fusion in the UL24syn background, suggesting that the two events may have an intrinsic link. Our results shed light on the biological properties of HSV-1 gI and may have important implications in understanding viral pathogenesis.
Keywords: cell-cell fusion; cell-to-cell spread; glycoprotein I (gI); herpes simplex virus; membrane tubulation; rod-shaped structures.
Copyright © 2020 American Society for Microbiology.
Figures

References
-
- Groves MJ. 2016. Genital herpes: a review. Am Fam Physician 93:928–934. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

