Rice GROWTH-REGULATING FACTOR7 Modulates Plant Architecture through Regulating GA and Indole-3-Acetic Acid Metabolism
- PMID: 32581114
- PMCID: PMC7479900
- DOI: 10.1104/pp.20.00302
Rice GROWTH-REGULATING FACTOR7 Modulates Plant Architecture through Regulating GA and Indole-3-Acetic Acid Metabolism
Abstract
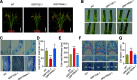
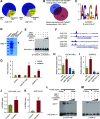
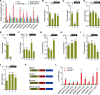
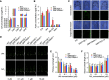
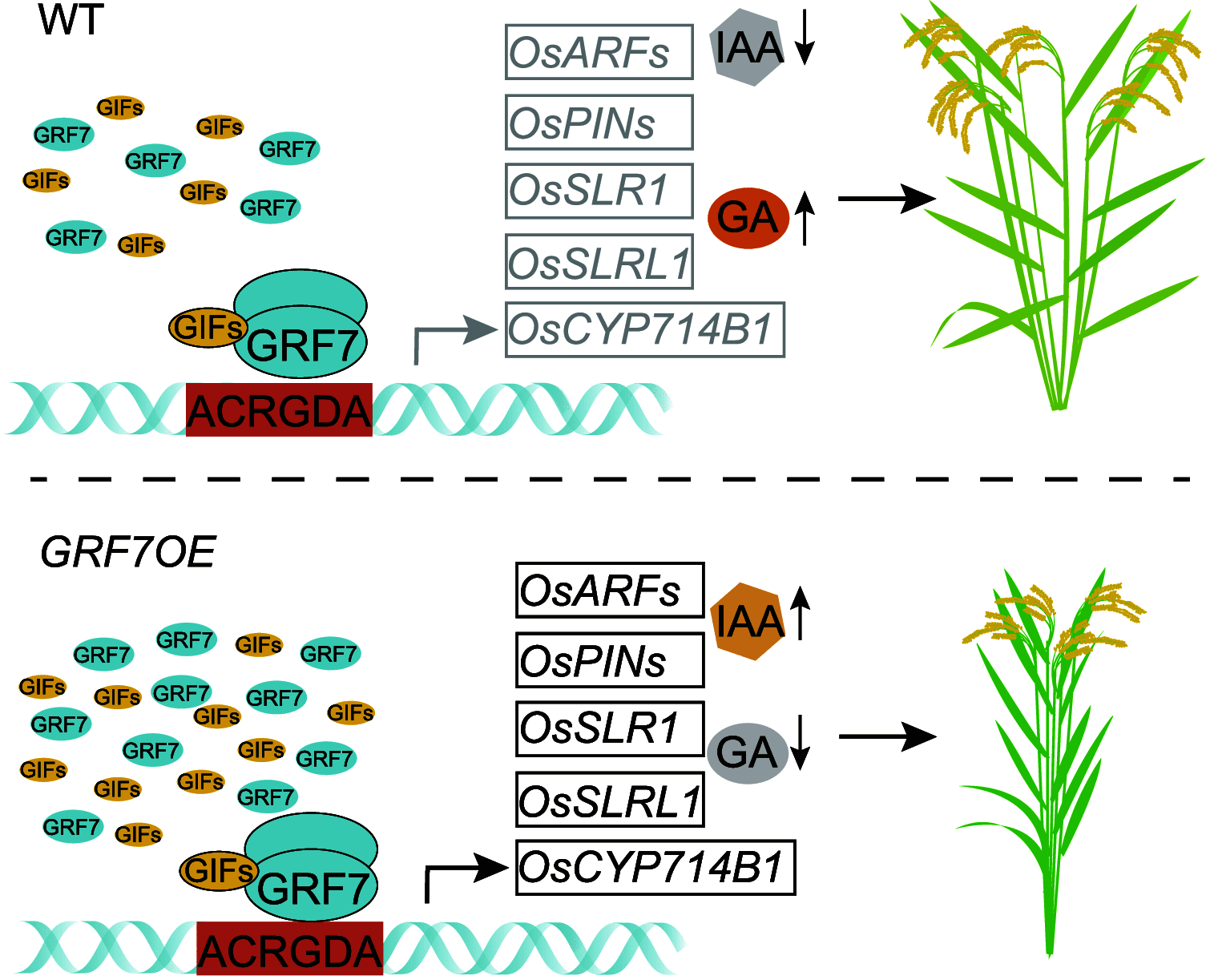
Plant-specific GROWTH-REGULATING FACTORs (GRFs) participate in central developmental processes, including leaf and root development; inflorescence, flower, and seed formation; senescence; and tolerance to stresses. In rice (Oryza sativa), there are 12 GRFs, but the role of the miR396-OsGRF7 regulatory module remains unknown. Here, we report that OsGRF7 shapes plant architecture via the regulation of auxin and GA metabolism in rice. OsGRF7 is mainly expressed in lamina joints, nodes, internodes, axillary buds, and young inflorescences. Overexpression of OsGRF7 causes a semidwarf and compact plant architecture with an increased culm wall thickness and narrowed leaf angles mediated by shortened cell length, altered cell arrangement, and increased parenchymal cell layers in the culm and adaxial side of the lamina joints. Knockout and knockdown lines of OsGRF7 exhibit contrasting phenotypes with severe degradation of parenchymal cells in the culm and lamina joints at maturity. Further analysis indicated that OsGRF7 binds the ACRGDA motif in the promoters of a cytochrome P450 gene and AUXIN RESPONSE FACTOR12, which are involved in the GA synthesis and auxin signaling pathways, respectively. Correspondingly, OsGRF7 alters the contents of endogenous GAs and auxins and sensitivity to exogenous phytohormones. These findings establish OsGRF7 as a crucial component in the OsmiR396-OsGRF-plant hormone regulatory network that controls rice plant architecture.
© 2020 American Society of Plant Biologists. All Rights Reserved.
Figures

References
-
- Baucher M, Moussawi J, Vandeputte OM, Monteyne D, Mol A, Pérez-Morga D, El Jaziri M(2013) A role for the miR396/GRF network in specification of organ type during flower development, as supported by ectopic expression of Populus trichocarpa miR396c in transgenic tobacco. Plant Biol (Stuttg) 15: 892–898 - PubMed
-
- Che R, Tong H, Shi B, Liu Y, Fang S, Liu D, Xiao Y, Hu B, Liu L, Wang H, et al. (2015) Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat Plants 2: 15195. - PubMed
-
- Chen X, Lu S, Wang Y, Zhang X, Lv B, Luo L, Xi D, Shen J, Ma H, Ming F(2015) OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice. Plant J 82: 302–314 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

