Human GBP1 binds LPS to initiate assembly of a caspase-4 activating platform on cytosolic bacteria
- PMID: 32581219
- PMCID: PMC7314798
- DOI: 10.1038/s41467-020-16889-z
Human GBP1 binds LPS to initiate assembly of a caspase-4 activating platform on cytosolic bacteria
Abstract
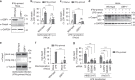
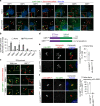
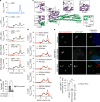
The human non-canonical inflammasome controls caspase-4 activation and gasdermin-D-dependent pyroptosis in response to cytosolic bacterial lipopolysaccharide (LPS). Since LPS binds and oligomerizes caspase-4, the pathway is thought to proceed without dedicated LPS sensors or an activation platform. Here we report that interferon-induced guanylate-binding proteins (GBPs) are required for non-canonical inflammasome activation by cytosolic Salmonella or upon cytosolic delivery of LPS. GBP1 associates with the surface of cytosolic Salmonella seconds after bacterial escape from their vacuole, initiating the recruitment of GBP2-4 to assemble a GBP coat. The GBP coat then promotes the recruitment of caspase-4 to the bacterial surface and caspase activation, in absence of bacteriolysis. Mechanistically, GBP1 binds LPS with high affinity through electrostatic interactions. Our findings indicate that in human epithelial cells GBP1 acts as a cytosolic LPS sensor and assembles a platform for caspase-4 recruitment and activation at LPS-containing membranes as the first step of non-canonical inflammasome signaling.
Conflict of interest statement
The authors declare no competing interests.
Figures




Comment in
-
Captain GBP1: inflammasomes assemble, pyroptotic endgame.Nat Immunol. 2020 Aug;21(8):829-830. doi: 10.1038/s41590-020-0727-0. Nat Immunol. 2020. PMID: 32699406 No abstract available.
References
-
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. - PubMed
-
- Kayagaki N, et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341:1246–1249. - PubMed
-
- Shi J, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

