Whole-genome sequencing of patients with rare diseases in a national health system
- PMID: 32581362
- PMCID: PMC7610553
- DOI: 10.1038/s41586-020-2434-2
Whole-genome sequencing of patients with rare diseases in a national health system
Abstract
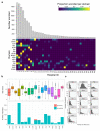
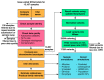
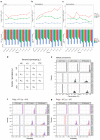
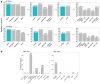
Most patients with rare diseases do not receive a molecular diagnosis and the aetiological variants and causative genes for more than half such disorders remain to be discovered1. Here we used whole-genome sequencing (WGS) in a national health system to streamline diagnosis and to discover unknown aetiological variants in the coding and non-coding regions of the genome. We generated WGS data for 13,037 participants, of whom 9,802 had a rare disease, and provided a genetic diagnosis to 1,138 of the 7,065 extensively phenotyped participants. We identified 95 Mendelian associations between genes and rare diseases, of which 11 have been discovered since 2015 and at least 79 are confirmed to be aetiological. By generating WGS data of UK Biobank participants2, we found that rare alleles can explain the presence of some individuals in the tails of a quantitative trait for red blood cells. Finally, we identified four novel non-coding variants that cause disease through the disruption of transcription of ARPC1B, GATA1, LRBA and MPL. Our study demonstrates a synergy by using WGS for diagnosis and aetiological discovery in routine healthcare.
Conflict of interest statement
Figures







References
-
- Ferreira CR. The burden of rare diseases. Am J Med Genet A. 2019 Jun;179(6):885–892. - PubMed
-
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015 May;17(5):405–24. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G1000848/MRC_/Medical Research Council/United Kingdom
- 202747/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- MR/S025502/1/MRC_/Medical Research Council/United Kingdom
- MR/K020919/1/MRC_/Medical Research Council/United Kingdom
- 201064/WT_/Wellcome Trust/United Kingdom
- MR/J011711/1/MRC_/Medical Research Council/United Kingdom
- 309548/ERC_/European Research Council/International
- MR/L006340/1/MRC_/Medical Research Council/United Kingdom
- 108749/WT_/Wellcome Trust/United Kingdom
- 29034/CRUK_/Cancer Research UK/United Kingdom
- MR/L019027/1/MRC_/Medical Research Council/United Kingdom
- MR/R002363/1/MRC_/Medical Research Council/United Kingdom
- 200990/WT_/Wellcome Trust/United Kingdom
- 204809/WT_/Wellcome Trust/United Kingdom
- FS/18/53/33863/BHF_/British Heart Foundation/United Kingdom
- MC_U120085815/MRC_/Medical Research Council/United Kingdom
- WT108749/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- G1002570/MRC_/Medical Research Council/United Kingdom
- MR/P02002X/1/MRC_/Medical Research Council/United Kingdom
- G0800571/MRC_/Medical Research Council/United Kingdom
- 107469/WT_/Wellcome Trust/United Kingdom
- SP/12/12/29836/BHF_/British Heart Foundation/United Kingdom
- 098519/WT_/Wellcome Trust/United Kingdom
- CH/1992001/6764/BHF_/British Heart Foundation/United Kingdom
- 202747/WT_/Wellcome Trust/United Kingdom
- RP-2016-07-019/DH_/Department of Health/United Kingdom
- G0701386/MRC_/Medical Research Council/United Kingdom
- 28051/CRUK_/Cancer Research UK/United Kingdom
- MC_PC_14089/MRC_/Medical Research Council/United Kingdom
- MC_PC_15080/MRC_/Medical Research Council/United Kingdom
- MR/M009203/1/MRC_/Medical Research Council/United Kingdom
- MR/N025431/2/MRC_/Medical Research Council/United Kingdom
- WT098519MA/WT_/Wellcome Trust/United Kingdom
- MR/R013942/1/MRC_/Medical Research Council/United Kingdom
- WT200990/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- 212219/WT_/Wellcome Trust/United Kingdom
- RG/16/4/32218/BHF_/British Heart Foundation/United Kingdom
- MC_EX_MR/M009203/1/MRC_/Medical Research Council/United Kingdom
- 107469/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- 109915/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- 090532/WT_/Wellcome Trust/United Kingdom
- 095908/WT_/Wellcome Trust/United Kingdom
- MR/N025431/1/MRC_/Medical Research Council/United Kingdom
- G1002528/MRC_/Medical Research Council/United Kingdom
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- FS/18/52/33808/BHF_/British Heart Foundation/United Kingdom
- 212219/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- MR/S021329/1/MRC_/Medical Research Council/United Kingdom
- 204809/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- MC_PC_18030/MRC_/Medical Research Council/United Kingdom
- MR/N010035/1/MRC_/Medical Research Council/United Kingdom
- 201064/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- MR/M012212/1/MRC_/Medical Research Council/United Kingdom
- 109915/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

