Chromosomal alterations among age-related haematopoietic clones in Japan
- PMID: 32581364
- PMCID: PMC7489641
- DOI: 10.1038/s41586-020-2426-2
Chromosomal alterations among age-related haematopoietic clones in Japan
Abstract
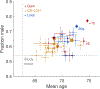
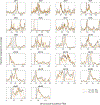
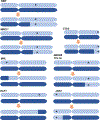
The extent to which the biology of oncogenesis and ageing are shaped by factors that distinguish human populations is unknown. Haematopoietic clones with acquired mutations become common with advancing age and can lead to blood cancers1-10. Here we describe shared and population-specific patterns of genomic mutations and clonal selection in haematopoietic cells on the basis of 33,250 autosomal mosaic chromosomal alterations that we detected in 179,417 Japanese participants in the BioBank Japan cohort and compared with analogous data from the UK Biobank. In this long-lived Japanese population, mosaic chromosomal alterations were detected in more than 35.0% (s.e.m., 1.4%) of individuals older than 90 years, which suggests that such clones trend towards inevitability with advancing age. Japanese and European individuals exhibited key differences in the genomic locations of mutations in their respective haematopoietic clones; these differences predicted the relative rates of chronic lymphocytic leukaemia (which is more common among European individuals) and T cell leukaemia (which is more common among Japanese individuals) in these populations. Three different mutational precursors of chronic lymphocytic leukaemia (including trisomy 12, loss of chromosomes 13q and 13q, and copy-neutral loss of heterozygosity) were between two and six times less common among Japanese individuals, which suggests that the Japanese and European populations differ in selective pressures on clones long before the development of clinically apparent chronic lymphocytic leukaemia. Japanese and British populations also exhibited very different rates of clones that arose from B and T cell lineages, which predicted the relative rates of B and T cell cancers in these populations. We identified six previously undescribed loci at which inherited variants predispose to mosaic chromosomal alterations that duplicate or remove the inherited risk alleles, including large-effect rare variants at NBN, MRE11 and CTU2 (odds ratio, 28-91). We suggest that selective pressures on clones are modulated by factors that are specific to human populations. Further genomic characterization of clonal selection and cancer in populations from around the world is therefore warranted.
Conflict of interest statement
No competing interests exist in this paper.
Figures

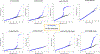




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

