Gene Deregulation and Underlying Mechanisms in Spinocerebellar Ataxias With Polyglutamine Expansion
- PMID: 32581696
- PMCID: PMC7296114
- DOI: 10.3389/fnins.2020.00571
Gene Deregulation and Underlying Mechanisms in Spinocerebellar Ataxias With Polyglutamine Expansion
Abstract
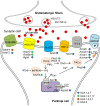
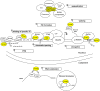
Polyglutamine spinocerebellar ataxias (polyQ SCAs) include SCA1, SCA2, SCA3, SCA6, SCA7, and SCA17 and constitute a group of adult onset neurodegenerative disorders caused by the expansion of a CAG repeat sequence located within the coding region of specific genes, which translates into polyglutamine tract in the corresponding proteins. PolyQ SCAs are characterized by degeneration of the cerebellum and its associated structures and lead to progressive ataxia and other diverse symptoms. In recent years, gene and epigenetic deregulations have been shown to play a critical role in the pathogenesis of polyQ SCAs. Here, we provide an overview of the functions of wild type and pathogenic polyQ SCA proteins in gene regulation, describe the extent and nature of gene expression changes and their pathological consequences in diseases, and discuss potential avenues to further investigate converging and distinct disease pathways and to develop therapeutic strategies.
Keywords: Purkinje cells; SCA; epigenetic; polyglutamine; spinocerebellar ataxia; transcriptional dysregulation.
Copyright © 2020 Niewiadomska-Cimicka, Hache and Trottier.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

