MiR-181d-5p Targets KLF6 to Improve Ischemia/Reperfusion-Induced AKI Through Effects on Renal Function, Apoptosis, and Inflammation
- PMID: 32581828
- PMCID: PMC7295155
- DOI: 10.3389/fphys.2020.00510
MiR-181d-5p Targets KLF6 to Improve Ischemia/Reperfusion-Induced AKI Through Effects on Renal Function, Apoptosis, and Inflammation
Abstract
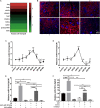
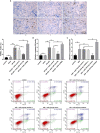
Renal tubular epithelial cell (RTEC) death and renal interstitial inflammation are the most crucial pathophysiological changes in acute kidney ischemia/reperfusion injury (IRI). The microRNA (miR)-181d family plays diverse roles in cell proliferation, apoptosis and inflammation, but its renal target and potential role in IRI are unknown. Here, we showed that the expression of miR-181d-5p decreased and Krueppel-like factor 6 (KLF6) increased in a renal cell (HK-2) model of hypoxia/reoxygenation (H/R) injury and a mouse model of renal IRI. They were mainly distributed in the renal tubules. After renal IRI, miR-181d-5p overexpression significantly inhibited inflammatory mediators, reduced apoptosis and further improved renal function. KLF6 exacerbated RTEC damage and acted as a NF-κB co-activator to aggravate the renal IRI inflammatory response. Mechanistically, KLF6 was predicted as a new potential target gene of miR-181d-5p through bioinformatic analysis and luciferase reporter assay verification. After overexpressing miR-181d-5p and inhibiting KLF6, the role of miR-181d-5p was weakened on the renal damage improvement. In conclusion, miR-181d-5p upregulation produced protective antiapoptotic and anti-inflammatory effects against IRI in kidneys in vivo and H/R injury in HK-2 cells in vitro, and these effects were achieved by targeted inhibition of KLF6. Thus, our results provide novel insights into the molecular mechanisms associated with IRI and a potential novel therapeutic target.
Keywords: IRI; KLF6; MiR-181d-5p; apoptosis; inflammation; renal function.
Copyright © 2020 Zhang, Li, Guan, Zhou, Wang, Yang, Zhen, Dai, Zhao, Jiang and Xu.
Figures






References
-
- Barnes P. J. (1997). Nuclear factor-kappa B. Int. J. Biochem. Cell Biol. 29 867–870. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

