Characterization and Genome Structure of Virulent Phage EspM4VN to Control Enterobacter sp. M4 Isolated From Plant Soft Rot
- PMID: 32582040
- PMCID: PMC7283392
- DOI: 10.3389/fmicb.2020.00885
Characterization and Genome Structure of Virulent Phage EspM4VN to Control Enterobacter sp. M4 Isolated From Plant Soft Rot
Abstract
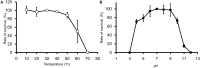
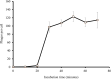
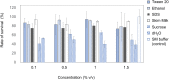
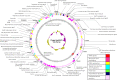
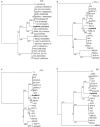
Enterobacter sp. M4 and other bacterial strains were isolated from plant soft rot disease. Virulent phages such as EspM4VN isolated from soil are trending biological controls for plant disease. This phage has an icosahedral head (100 nm in diameter), a neck, and a contractile sheath (100 nm long and 18 nm wide). It belongs to the Ackermannviridae family and resembles Shigella phage Ag3 and Dickeya phages JA15 and XF4. We report herein that EspM4VN was stable from 10°C to 50°C and pH 4 to 10 but deactivated at 70°C and pH 3 and 12. This phage formed clear plaques only on Enterobacter sp. M4 among tested bacterial strains. A one-step growth curve showed that the latent phase was 20 min, rise period was 10 min, and an average of 122 phage particles were released from each absorbed cell. We found the phage's genome size was 160,766 bp and that it annotated 219 open reading frames. The genome organization of EspM4VN has high similarity with the Salmonella phage SKML-39; Dickeya phages Coodle, PP35, JA15, and Limestone; and Shigella phage Ag3. The phage EspM4VN has five tRNA species, four tail-spike proteins, and a thymidylate synthase. Phylogenetic analysis based on structural proteins and enzymes indicated that EspM4VN was identified as a member of the genus Agtrevirus, subfamily Aglimvirinae, family Ackermannviridae.
Keywords: Agtrevirus; Enterobacter; genome; soft rot disease; virulent phage.
Copyright © 2020 Thanh, Nagayoshi, Fujino, Iiyama, Furuya, Hiromasa, Iwamoto and Doi.
Figures



References
-
- Adams M. H. (1959). Bacteriophages. New York, NY: Interscience Publishers Inc.
-
- Adeolu M., Alnajar S., Naushad S., Gupta R. S. (2016). Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 66 5575–5599. 10.1099/ijsem.0.001485 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

