Cloning, Expression, and Structural Elucidation of a Biotechnologically Potential Alkaline Serine Protease From a Newly Isolated Haloalkaliphilic Bacillus lehensis JO-26
- PMID: 32582046
- PMCID: PMC7283590
- DOI: 10.3389/fmicb.2020.00941
Cloning, Expression, and Structural Elucidation of a Biotechnologically Potential Alkaline Serine Protease From a Newly Isolated Haloalkaliphilic Bacillus lehensis JO-26
Abstract
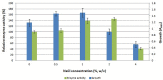
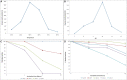
An alkaline protease gene of Bacillus lehensis JO-26 from saline desert, Little Rann of Kutch, was cloned and expressed in Escherichia coli BL21 (DE3). A 1,014-bp ORF encoded 337 amino acids. The recombinant protease (APrBL) with Asp 97, His 127, and Ser 280 forming catalytic triad belongs to the subtilase S8 protease family. The gene was optimally expressed in soluble fraction with 0.2 mM isopropyl β-D-thiogalactopyranoside (IPTG), 2% (w/v) NaCl at 28°C. APrBL, a monomer with a molecular mass of 34.6 kDa was active over pH 8-11 and 30°C-70°C, optimally at pH 10 and 50°C. The enzyme was highly thermostable and retained 73% of the residual activity at 80°C up to 3 h. It was significantly stimulated by sodium dodecyl sulfate (SDS), Ca2+, chloroform, toluene, n-butanol, and benzene while completely inhibited by phenylmethylsulfonyl fluoride (PMSF) and Hg2+. The serine nature of the protease was confirmed by its strong inhibition by PMSF. The APrBL gene was phylogenetically close to alkaline elastase YaB (P20724) and was distinct from the well-known commercial proteases subtilisin Carlsberg (CAB56500) and subtilisin BPN' (P00782). The structural elucidation revealed 31.75% α-helices, 22.55% β-strands, and 45.70% coils. Although high glycine and fewer proline residues are a characteristic feature of the cold-adapted enzymes, the similar observation in thermally active APrBL suggests that this feature cannot be solely responsible for thermo/cold adaptation. The APrBL protease was highly effective as a detergent additive and in whey protein hydrolysis.
Keywords: Little Rann of Kutch; detergent additive; gene expression; recombinant alkaline protease; structure–function relationship; whey protein hydrolysis.
Copyright © 2020 Bhatt and Singh.
Figures





References
-
- Annamalai N., Rajeswari M. V., Balasubramanian T. (2014). Extraction, purification and application of thermostable and halostable alkaline protease from Bacillus alveayuensis CAS 5 using marine wastes. Food Bioprod Process 92 335–342. 10.1016/j.fbp.2013.08.009 - DOI
-
- Barberis S., Quiroga E., Morcelle S., Priolo N., Luco J. M. (2006). Study of phytoproteases stability in aqueous-organic biphasic systems using linear free energy relationships. J. Mol. Catal. 38 95–103. 10.1016/j.molcatb.2005.11.011 - DOI
-
- Beg Q. K., Gupta R. (2003). Purification and characterization of an oxidation-stable, thiol-dependent serine alkaline protease from Bacillus mojavensis. Enzyme Microb. Technol. 32 294–304. 10.1016/S0141-0229(02)00293-4 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

