The Gene Catalog and Comparative Analysis of Gut Microbiome of Big Cats Provide New Insights on Panthera Species
- PMID: 32582053
- PMCID: PMC7287027
- DOI: 10.3389/fmicb.2020.01012
The Gene Catalog and Comparative Analysis of Gut Microbiome of Big Cats Provide New Insights on Panthera Species
Abstract
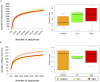
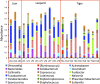
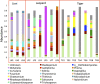
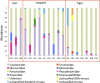
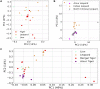
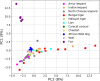
Majority of metagenomic studies in the last decade have focused on revealing the gut microbiomes of humans, rodents, and ruminants; however, the gut microbiome and genic information (gene catalog) of large felids such as Panthera species are largely unknown to date. In this study, the gut bacterial, fungal, and viral metagenomic composition was assessed from three Panthera species (lion, leopard, and tiger) of Indian origin, which were consuming the same diet and belonged to the same geographical location. A non-redundant bacterial gene catalog of the Panthera gut consisting of 1,507,035 putative genes was constructed from 27 Panthera individuals, which revealed a higher abundance of purine metabolism genes correlating with their purine-rich dietary intake. Analysis with Carbohydrate Active enZyme (CAZy) and MEROPS databases identified enrichment of glycoside hydrolases (GHs), glycoside-transferases, and collagenases in the gut, which are important for nutrient acquisition from animal biomass. The bacterial, fungal, and viral community analysis provided the first comprehensive insights into the Panthera-specific microbial community. The Panthera gene catalog and the largest comparative study of the gut bacterial composition of 68 individuals of Carnivora species from different geographical locations and diet underscore the role of diet and geography in shaping the Panthera gut microbiome, which is significant for the health and conservation management of these highly endangered species.
Keywords: Indian Panthera; big cats; gut microbiome; hypercarnivores; metagenomics.
Copyright © 2020 Mittal, Saxena, Gupta, Mahajan and Sharma.
Figures

References
LinkOut - more resources
Full Text Sources
Miscellaneous

