Special Features of Bat Microbiota Differ From Those of Terrestrial Mammals
- PMID: 32582057
- PMCID: PMC7284282
- DOI: 10.3389/fmicb.2020.01040
Special Features of Bat Microbiota Differ From Those of Terrestrial Mammals
Abstract
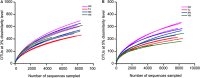
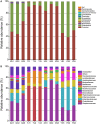
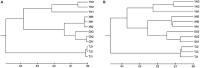
Bats (order Chiroptera) are one of the most diverse and widely distributed group of mammals with a close relationship to humans. Over the past few decades, a number of studies have been performed on bat viruses; in contrast, bacterial pathogens carried by bats were largely neglected. As more bacterial pathogens are being identified from bats, the need to study their natural microbiota is becoming urgent. In the current study, fecal samples of four bat species from different locations of China were analyzed for their microbiota composition. Together with the results of others, we concluded that bat microbiota is most commonly dominated by Firmicutes and Proteobacteria; the strict anaerobic phylum Bacteroidetes, which is dominant in other terrestrial mammals, especially humans and mice, is relatively rare in bats. This phenomenon was interpreted as a result of a highly specified gastrointestinal tract in adaptation to the flying lifestyle of bats. Further comparative study implied that bat microbiota resemble those of the order Carnivora. To discover potential bacterial pathogens, a database was generated containing the 16S rRNA gene sequences of known bacterial pathogens. Potential bacterial pathogens belonging to 12 genera were detected such as Salmonella, Shigella, and Yersinia, among which some have been previously reported in bats. This study demonstrated high resolution and repeatability in detecting organisms of rare existence, and the results could be used as guidance for future bacterial pathogen isolation.
Keywords: aerobic microbes; bacterial pathogen; bat; gut microbiota; sequencing.
Copyright © 2020 Sun, Gao, Ge, Shi and Zhou.
Figures



References
-
- Arata A. A., Vaughn J. B., Newell K. W., Barth R. A. J., Gracian M. (1968). Salmonella and Shigella infections in bats in selected areas of Colombia. Am. J. Trop. Med. Hyg. 17 92–95. - PubMed
-
- Canals M., Atala C., Olivares R., Guajardo F., Figueroa D. P., Sabat P., et al. (2005). Functional and structural optimization of the respiratory system of the bat Tadarida brasiliensis (Chiroptera, Molossidae): does airway geometry matter? J. Exp. Biol. 208 3987–3995. 10.1242/jeb.01817 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

