Bioprospecting Microbial Diversity for Lignin Valorization: Dry and Wet Screening Methods
- PMID: 32582068
- PMCID: PMC7295907
- DOI: 10.3389/fmicb.2020.01081
Bioprospecting Microbial Diversity for Lignin Valorization: Dry and Wet Screening Methods
Abstract
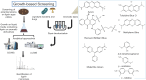
Lignin is an abundant cell wall component, and it has been used mainly for generating steam and electricity. Nevertheless, lignin valorization, i.e. the conversion of lignin into high value-added fuels, chemicals, or materials, is crucial for the full implementation of cost-effective lignocellulosic biorefineries. From this perspective, rapid screening methods are crucial for time- and resource-efficient development of novel microbial strains and enzymes with applications in the lignin biorefinery. The present review gives an overview of recent developments and applications of a vast arsenal of activity and sequence-based methodologies for uncovering novel microbial strains with ligninolytic potential, novel enzymes for lignin depolymerization and for unraveling the main metabolic routes during growth on lignin. Finally, perspectives on the use of each of the presented methods and their respective advantages and disadvantages are discussed.
Keywords: activity-based screening; bioprocess; biosensors; high-throughput screening; lignin-degrading enzymes.
Copyright © 2020 Gonçalves, Bruce, Silva, Fillho, Noronha, Carlquist and Parachin.
Figures





References
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

