Characterization of a Novel Functional Trimeric Catechol 1,2-Dioxygenase From a Pseudomonas stutzeri Isolated From the Gulf of Mexico
- PMID: 32582076
- PMCID: PMC7287156
- DOI: 10.3389/fmicb.2020.01100
Characterization of a Novel Functional Trimeric Catechol 1,2-Dioxygenase From a Pseudomonas stutzeri Isolated From the Gulf of Mexico
Abstract
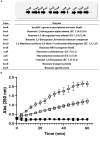
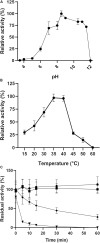
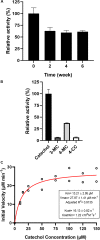
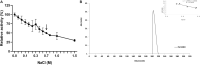
Catechol 1,2 dioxygenases (C12DOs) have been studied for its ability to cleavage the benzene ring of catechol, the main intermediate in the degradation of aromatic compounds derived from aerobic degradation of hydrocarbons. Here we report the genome sequence of the marine bacterium Pseudomonas stutzeri GOM2, isolated from the southwestern Gulf of Mexico, and the biochemical characterization of its C12DO (PsC12DO). The catA gene, encoding PsC12DO of 312 amino acid residues, was cloned and expressed in Escherichia coli. Many C12DOs have been described as dimeric enzymes including those present in Pseudomonas species. The purified PsC12DO enzyme was found as an active trimer, with a molecular mass of 107 kDa. Increasing NaCl concentration in the enzyme reaction gradually reduced activity; in high salt concentrations (0.7 M NaCl) quaternary structural analysis determined that the enzyme changes to a dimeric arrangement and causes a 51% decrease in specific activity on catechol substrate. In comparison with other C12DOs, our enzyme showed a broad range of action for PsC12DO in solutions with pH values ranging from neutral to alkaline (70%). The enzyme is still active after incubation at 50°C for 30 min and in low temperatures to long term storage after 6 weeks at 4°C (61%). EDTA or Ca2+ inhibitors cause no drastic changes on residual activity; nevertheless, the activity of the enzyme was affected by metal ions Fe3+, Zn2+ and was completely inhibited by Hg2+. Under optimal conditions the k cat and K m values were 16.13 s-1 and 13.2 μM, respectively. To our knowledge, this is the first report describing the characterization of a marine C12DOs from P. stutzeri isolated from the Gulf of Mexico that is active in a trimeric state. We consider that our enzyme has important features to be used in environments in presence of EDTA, metals and salinity conditions.
Keywords: Pseudomonas; aromatic compounds; catechol degradation; intradiol dioxygenase; trimeric structure.
Copyright © 2020 Rodríguez-Salazar, Almeida-Juarez, Ornelas-Ocampo, Millán-López, Raga-Carbajal, Rodríguez-Mejía, Muriel-Millán, Godoy-Lozano, Rivera-Gómez, Rudiño-Piñera and Pardo-López.
Figures




References
-
- Al-Hakim M. H., Hasan R., Ali M. H., Rabbee M. F., Marufatuzzahan H. M., Joy Z. F. (2015). In-silico characterization and homology modeling of catechol 1,2 dioxygenase involved in processing of catechol- an intermediate of aromatic compound degradation pathway. Glob. J. Sci. Front. Res. G Bio-Tech Genet. 15 1–13.
-
- Bagdasarian M., Lurz R., Ruckert B., Franklin F. C., Bagdasarian M. M., Frey J., et al. (1981). Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16 237–247. 10.1016/0378-1119(81)90080-9 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

