Growth and Break-Up of Methanogenic Granules Suggests Mechanisms for Biofilm and Community Development
- PMID: 32582085
- PMCID: PMC7285868
- DOI: 10.3389/fmicb.2020.01126
Growth and Break-Up of Methanogenic Granules Suggests Mechanisms for Biofilm and Community Development
Abstract
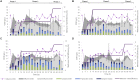
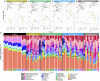
Methanogenic sludge granules are densely packed, small, spherical biofilms found in anaerobic digesters used to treat industrial wastewaters, where they underpin efficient organic waste conversion and biogas production. Each granule theoretically houses representative microorganisms from all of the trophic groups implicated in the successive and interdependent reactions of the anaerobic digestion (AD) process. Information on exactly how methanogenic granules develop, and their eventual fate will be important for precision management of environmental biotechnologies. Granules from a full-scale bioreactor were size-separated into small (0.6-1 mm), medium (1-1.4 mm), and large (1.4-1.8 mm) size fractions. Twelve laboratory-scale bioreactors were operated using either small, medium, or large granules, or unfractionated sludge. After >50 days of operation, the granule size distribution in each of the small, medium, and large bioreactor sets had diversified beyond-to both bigger and smaller than-the size fraction used for inoculation. Interestingly, extra-small (XS; <0.6 mm) granules were observed, and retained in all of the bioreactors, suggesting the continuous nature of granulation, and/or the breakage of larger granules into XS bits. Moreover, evidence suggested that even granules with small diameters could break. "New" granules from each emerging size were analyzed by studying community structure based on high-throughput 16S rRNA gene sequencing. Methanobacterium, Aminobacterium, Propionibacteriaceae, and Desulfovibrio represented the majority of the community in new granules. H2-using, and not acetoclastic, methanogens appeared more important, and were associated with abundant syntrophic bacteria. Multivariate integration (MINT) analyses identified distinct discriminant taxa responsible for shaping the microbial communities in different-sized granules.
Keywords: anaerobic digestion; biofilms; methanogens; microbial communities; sludge granules; wastewater.
Copyright © 2020 Trego, Galvin, Sweeney, Dunning, Murphy, Mills, Nzeteu, Quince, Connelly, Ijaz and Collins.
Figures



References
-
- Afridi Z. U. R., Wu J., Cao Z. P., Zhang Z. L., Li Z. H., Poncin S., et al. (2017). Insight into mass transfer by convective diffusion in anaerobic granules to enhance biogas production. Biochem. Eng. J. 127 154–160. 10.1016/j.bej.2017.07.012 - DOI
-
- Ahn J.-H., Forster C. F. (2000). Kinetic analyses of the operation of mesophilic and thermophilic anaerobic filters treating a simulated starch wastewater. Process Biochem. 36 19–23. 10.1016/S0032-9592(00)00166-7 - DOI
-
- Ahn Y. (2000). Physicochemical and microbial aspects of anaerobic granular biopellets. J. Environ. Sci. Health A 35 1617–1635. 10.1080/10934520009377059 - DOI
-
- Akasaka H., Ueki A., Hanada S., Kamagata Y., Ueki K. (2003). Propionicimonas paludicola gen. nov., sp. nov., a novel facultatively anaerobic, Gram-positive, propionate-producing bacterium isolated from plant residue in irrigated rice-field soil. Int. J. Syst. Evol. Microbiol. 53 1991–1998. 10.1099/ijs.0.02764-0 - DOI - PubMed
-
- Arcand Y., Guiot S. R., Desrochers M., Chavarie C. (1994). Impact of the reactor hydrodynamics and organic loading on the size and activity of anaerobic granules. Chem. Eng. J. Biochem. Eng. J. 56 B23–B35. 10.1016/0923-0467(94)87028-4 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

