Identification of Microbial Genetic Capacities and Potential Mechanisms Within the Rumen Microbiome Explaining Differences in Beef Cattle Feed Efficiency
- PMID: 32582125
- PMCID: PMC7292206
- DOI: 10.3389/fmicb.2020.01229
Identification of Microbial Genetic Capacities and Potential Mechanisms Within the Rumen Microbiome Explaining Differences in Beef Cattle Feed Efficiency
Abstract
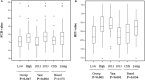
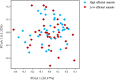
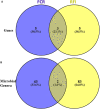
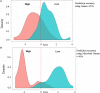
In this study, Bos Taurus cattle offered one high concentrate diet (92% concentrate-8% straw) during two independent trials allowed us to classify 72 animals comprising of two cattle breeds as "Low" or "High" feed efficiency groups. Digesta samples were taken from individual beef cattle at the abattoir. After metagenomic sequencing, the rumen microbiome composition and genes were determined. Applying a targeted approach based on current biological evidence, 27 genes associated with host-microbiome interaction activities were selected. Partial least square analysis enabled the identification of the most significant genes and genera of feed efficiency (VIP > 0.8) across years of the trial and breeds when comparing all potential genes or genera together. As a result, limited number of genes explained about 40% of the variability in both feed efficiency indicators. Combining information from rumen metagenome-assembled genomes and partial least square analysis results, microbial genera carrying these genes were determined and indicated that a limited number of important genera impacting on feed efficiency. In addition, potential mechanisms explaining significant difference between Low and High feed efficiency animals were analyzed considering, based on the literature, their gastrointestinal location of action. High feed efficiency animals were associated with microbial species including several Eubacterium having the genetic capacity to form biofilm or releasing metabolites like butyrate or propionate known to provide a greater contribution to cattle energy requirements compared to acetate. Populations associated with fucose sensing or hemolysin production, both mechanisms specifically described in the lower gut by activating the immune system to compete with pathogenic colonizers, were also identified to affect feed efficiency using rumen microbiome information. Microbial mechanisms associated with low feed efficiency animals involved potential pathogens within Proteobacteria and Spirochaetales, releasing less energetic substrates (e.g., acetate) or producing sialic acid to avoid the host immune system. Therefore, this study focusing on genes known to be involved in host-microbiome interaction improved the identification of rumen microbial genetic capacities and potential mechanisms significantly impacting on feed efficiency in beef cattle fed high concentrate diet.
Keywords: feed conversion efficiency; high concentrate diet; metagenomic sequencing; predicted microbial mechanisms; rumen microbiome.
Copyright © 2020 Auffret, Stewart, Dewhurst, Duthie, Watson and Roehe.
Figures
References
-
- Abt B., Han C., Scheuner C., Lu M., Lapidus A., Nolan M., et al. (2012). Complete genome sequence of the termite hindgut bacterium Spirochaeta coccoides type strain (SPN1T), reclassification in the genus Sphaerochaeta as Sphaerochaeta coccoides comb. nov. and emendations of the family Spirochaetaceae and the genus Sphaerochaeta. Stand. Genomic Sci. 6 194–209. 10.4056/sigs.2796069 - DOI - PMC - PubMed
-
- Auffret M. D., Stewart R., Dewhurst R. J., Duthie C.-A., Rooke J. A., Wallace R. J., et al. (2018). Identification, comparison, and validation of robust rumen microbial biomarkers for methane emissions using diverse Bos taurus breeds and basal diets. Front. Microbiol. 8:2642. 10.3389/fmicb.2017.02642 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

