Genomic Origin and Diversification of the Glucosinolate MAM Locus
- PMID: 32582245
- PMCID: PMC7289053
- DOI: 10.3389/fpls.2020.00711
Genomic Origin and Diversification of the Glucosinolate MAM Locus
Abstract
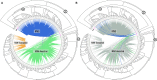
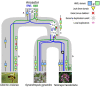
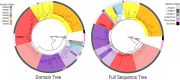
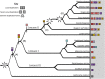
Glucosinolates are a diverse group of plant metabolites that characterize the order Brassicales. The MAM locus is one of the most significant QTLs for glucosinolate diversity. However, most of what we understand about evolution at the locus is focused on only a few species and not within a phylogenetic context. In this study, we utilize a micro-synteny network and phylogenetic inference to investigate the origin and diversification of the MAM/IPMS gene family. We uncover unique MAM-like genes found at the orthologous locus in the Cleomaceae that shed light on the transition from IPMS to MAM. In the Brassicaceae, we identify six distinct MAM clades across Lineages I, II, and III. We characterize the evolutionary impact and consequences of local duplications, transpositions, whole genome duplications, and gene fusion events, generating several new hypothesizes on the function and diversity of the MAM locus.
Keywords: brassicaceae; gene duplication; gene family; gene fusion; glucosinolates; polyploidy.
Copyright © 2020 Abrahams, Pires and Schranz.
Figures
References
-
- Benderoth M., Pfalz M., Kroymann J. (2009). Methylthioalkylmalate synthases: genetics, ecology and evolution. Phytochem. Rev. 8 255–268. 10.1007/s11101-008-9097-1 - DOI
LinkOut - more resources
Full Text Sources

