Bioinformatics Analysis of Prognostic miRNA Signature and Potential Critical Genes in Colon Cancer
- PMID: 32582275
- PMCID: PMC7296168
- DOI: 10.3389/fgene.2020.00478
Bioinformatics Analysis of Prognostic miRNA Signature and Potential Critical Genes in Colon Cancer
Abstract
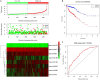
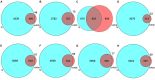
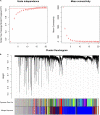
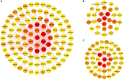
This study aims to lay a foundation for studying the regulation of microRNAs (miRNAs) in colon cancer by applying bioinformatics methods to identify miRNAs and their potential critical target genes associated with colon cancer and prognosis. Data of differentially expressed miRNAs (DEMs) and genes (DEGs) downloaded from two independent databases (TCGA and GEO) and analyzed by R software resulted in 472 DEMs and 565 DEGs in colon cancers, respectively. Next, we developed an 8-miRNA (hsa-mir-6854, hsa-mir-4437, hsa-mir-216a, hsa-mir-3677, hsa-mir-887, hsa-mir-4999, hsa-mir-34b, and hsa-mir-3189) prognostic signature for patients with colon cancer by Cox proportional hazards regression analysis. To predict the target genes of these miRNAs, we used TargetScan and miRDB. The intersection of DEGs with the target genes predicted for these eight miRNAs retrieved 112 consensus genes. GO and KEGG pathway enrichment analyses showed these 112 genes were mainly involved in protein binding, one-carbon metabolic process, nitrogen metabolism, proteoglycans in cancer, and chemokine signaling pathways. The protein-protein interaction network of the consensus genes, constructed using the STRING database and imported into Cytoscape, identified 14 critical genes in the pathogenesis of colon cancer (CEP55, DTL, FANCI, HMMR, KIF15, MCM6, MKI67, NCAPG2, NEK2, RACGAP1, RRM2, TOP2A, UBE2C, and ZWILCH). Finally, we verified the critical genes by weighted gene co-expression network analysis (WGCNA) of the GEO data, and further mined the core genes involved in colon cancer. In summary, this study identified an 8-miRNA model that can effectively predict the prognosis of colon cancer patients and 14 critical genes with vital roles in colon cancer carcinogenesis. Our findings contribute new ideas for elucidating the molecular mechanisms of colon cancer carcinogenesis and provide new therapeutic targets and biomarkers for future treatment and prognosis.
Keywords: GEO; TCGA; bioinformatics; biomarker; colon cancer; microRNA; prognosis.
Copyright © 2020 Chen, Gao, Liu, Wen, Hong and Huang.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

