Analysis of Circular RNA-Related Competing Endogenous RNA Identifies the Immune-Related Risk Signature for Colorectal Cancer
- PMID: 32582276
- PMCID: PMC7283524
- DOI: 10.3389/fgene.2020.00505
Analysis of Circular RNA-Related Competing Endogenous RNA Identifies the Immune-Related Risk Signature for Colorectal Cancer
Abstract
Background: Recent papers have described circular RNAs (circRNAs) playing important roles in the development and progression of colorectal cancer (CRC). However, the expression profiles of circRNAs and their functions in CRC have rarely been studied. The objective was to identify circRNAs involved in the carcinogenesis and progression of CRC and to explore potential molecular mechanisms as a competitive endogenous RNA (ceRNA). Moreover, we aimed to establish an immune-related gene signature for predicting the overall survival (OS) of CRC.
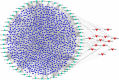
Methods: The expression patterns of circRNA, miRNA, mRNA, and clinicopathological data were collected from the GEO and TCGA databases. A ceRNA network would be established, and the functional enrichment analyses were performed. The protein-protein interaction network (PPI) was constructed, and hub genes were identified using a cytohub plugin. Subsequently, an immune-related signature was developed based on mRNAs in the ceRNA network. In addition, OS-nomogram was constructed by combining an immune-related signature and clinicopathological characterization to predict the OS.
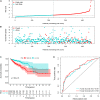
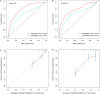
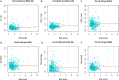
Results: We established a circRNA-miRNA-mRNA ceRNA network. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed that the mRNAs were mainly enriched in neuroactive ligand-receptor interaction, Wnt signaling pathway, cell adhesion molecules (CAMs), and renin secretion. PPI network and module analysis identified 10 hub genes, and the circRNA-miRNA hub gene regulatory modules was established. After univariate and multivariate analysis, seven immune-related genes in the ceRNA network were used to construct the immune-related signature. Patients were divided into low-risk and high-risk groups, and there were significant differences in the OS. The ROC of the nomogram indicated the satisfactory accuracy and predictive power. Furthermore, we established a prognostic nomogram based on immune-related risk score and clinical characterization. The ROC and calibration curves revealed the accuracy of the nomogram. In addition, the high-risk score was positively correlated with six immune infiltrating cells (P < 0.05).
Conclusion: We screened the key genes and established a circRNA-related ceRNA network involved in CRC, which will assist in understanding the molecular mechanisms underlying the carcinogenesis and progression. Moreover, our proposed immune-based signature may predict survival and reflect the immune status of CRC patients.
Keywords: circRNA; colorectal cancer; competitive endogenous RNA; immune-related genes; prognostic signature.
Copyright © 2020 Song, Ren, Wang, Ge and Fu.
Figures




References
-
- Amin M. B., Greene F. L., Edge S. B., Compton C. C., Gershenwald J. E., Brookland R. K., et al. (2017). The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 67 93–99. 10.3322/caac.21388 - DOI - PubMed
LinkOut - more resources
Full Text Sources

