Improving Industrially Relevant Phenotypic Traits by Engineering Chromosome Copy Number in Saccharomyces pastorianus
- PMID: 32582279
- PMCID: PMC7283523
- DOI: 10.3389/fgene.2020.00518
Improving Industrially Relevant Phenotypic Traits by Engineering Chromosome Copy Number in Saccharomyces pastorianus
Abstract
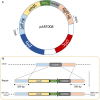
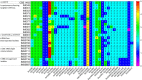
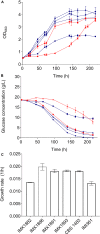
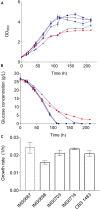
The lager-brewing yeast Saccharomyces pastorianus is a hybrid between S. cerevisiae and S. eubayanus with an exceptional degree of aneuploidy. While chromosome copy number variation (CCNV) is present in many industrial Saccharomyces strains and has been linked to various industrially-relevant traits, its impact on the brewing performance of S. pastorianus remains elusive. Here we attempt to delete single copies of chromosomes which are relevant for the production of off-flavor compound diacetyl by centromere silencing. However, the engineered strains display CNV of multiple non-targeted chromosomes. We attribute this unintended CCNV to inherent instability and to a mutagenic effect of electroporation and of centromere-silencing. Regardless, the resulting strains displayed large phenotypic diversity. By growing centromere-silenced cells in repeated sequential batches in medium containing 10% ethanol, mutants with increased ethanol tolerance were obtained. By using CCNV mutagenesis by exposure to the mitotic inhibitor MBC, selection in the same set-up yielded even more tolerant mutants that would not classify as genetically modified organisms. These results show that CCNV of alloaneuploid S. pastorianus genomes is highly unstable, and that CCNV mutagenesis can generate broad diversity. Coupled to effective selection or screening, CCNV mutagenesis presents a potent tool for strain improvement.
Keywords: Saccharomyces pastorianus; chromosome copy number stability; chromosome missegregation; lager beer brewing; strain engineering.
Copyright © 2020 Gorter de Vries, Knibbe, van Roosmalen, van den Broek, de la Torre Cortés, O’Herne, Vijverberg, el Masoudi, Brouwers, Pronk and Daran.
Figures




Similar articles
-
Chromosomal Copy Number Variation in Saccharomyces pastorianus Is Evidence for Extensive Genome Dynamics in Industrial Lager Brewing Strains.Appl Environ Microbiol. 2015 Sep;81(18):6253-67. doi: 10.1128/AEM.01263-15. Epub 2015 Jul 6. Appl Environ Microbiol. 2015. PMID: 26150454 Free PMC article.
-
Industrially Applicable De Novo Lager Yeast Hybrids with a Unique Genomic Architecture: Creation and Characterization.Appl Environ Microbiol. 2021 Jan 15;87(3):e02434-20. doi: 10.1128/AEM.02434-20. Print 2021 Jan 15. Appl Environ Microbiol. 2021. PMID: 33188002 Free PMC article.
-
Himalayan Saccharomyces eubayanus Genome Sequences Reveal Genetic Markers Explaining Heterotic Maltotriose Consumption by Saccharomyces pastorianus Hybrids.Appl Environ Microbiol. 2019 Oct 30;85(22):e01516-19. doi: 10.1128/AEM.01516-19. Print 2019 Nov 15. Appl Environ Microbiol. 2019. PMID: 31519660 Free PMC article.
-
Industrial Relevance of Chromosomal Copy Number Variation in Saccharomyces Yeasts.Appl Environ Microbiol. 2017 May 17;83(11):e03206-16. doi: 10.1128/AEM.03206-16. Print 2017 Jun 1. Appl Environ Microbiol. 2017. PMID: 28341679 Free PMC article. Review.
-
Saccharomyces pastorianus: genomic insights inspiring innovation for industry.Yeast. 2015 Jan;32(1):17-27. doi: 10.1002/yea.3033. Epub 2014 Sep 23. Yeast. 2015. PMID: 25088523 Review.
Cited by
-
Spontaneous and environment induced genomic alterations in yeast model.Cell Insight. 2024 Sep 26;4(1):100209. doi: 10.1016/j.cellin.2024.100209. eCollection 2025 Feb. Cell Insight. 2024. PMID: 39629481 Free PMC article. Review.
-
Genomic instability in an interspecific hybrid of the genus Saccharomyces: a matter of adaptability.Microb Genom. 2020 Oct;6(10):mgen000448. doi: 10.1099/mgen.0.000448. Microb Genom. 2020. PMID: 33021926 Free PMC article.
-
Transcriptional Profile of the Industrial Hybrid Saccharomyces pastorianus Reveals Temperature-Dependent Allele Expression Bias and Preferential Orthologous Protein Assemblies.Mol Biol Evol. 2021 Dec 9;38(12):5437-5452. doi: 10.1093/molbev/msab282. Mol Biol Evol. 2021. PMID: 34550394 Free PMC article.
References
-
- Bolat I., Romagnoli G., Zhu F., Pronk J. T., Daran J. M. (2013). Functional analysis and transcriptional regulation of two orthologs of ARO10, encoding broad-substrate-specificity 2-oxo-acid decarboxylases, in the brewing yeast Saccharomyces pastorianus CBS 1483. FEMS Yeast Res. 13 505–517. 10.1111/1567-1364.12051 - DOI - PubMed
-
- Bolat I., Walsh M. C., Turtoi M. (2008). Isolation and characterization of two lager yeast strains from the WS34/70 population. Roum. Biotechnol. Lett. 13 62–73.
-
- Bracher J. M., de Hulster E., Koster C. C., van den Broek M., Daran J. G., van Maris A. J. A., et al. (2017). Laboratory evolution of a biotin-requiring Saccharomyces cerevisiae strain for full biotin prototrophy and identification of causal mutations. Appl. Environ. Microbiol. 83:e00892-17. 10.1128/AEM.00892-17 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

