Ancestral Haplotype Mapping for GWAS and Detection of Signatures of Selection in Admixed Dairy Cattle of Kenya
- PMID: 32582285
- PMCID: PMC7296079
- DOI: 10.3389/fgene.2020.00544
Ancestral Haplotype Mapping for GWAS and Detection of Signatures of Selection in Admixed Dairy Cattle of Kenya
Abstract
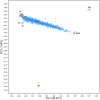
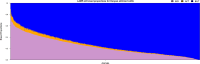
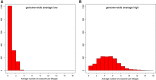
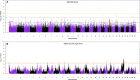
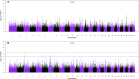
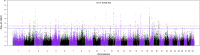
Understanding the genetic structure of adaptation and productivity in challenging environments is necessary for designing breeding programs that suit such conditions. Crossbred dairy cattle in East Africa resulting from over 60 years of crossing exotic dairy breeds with indigenous cattle plus inter se matings form a highly variable admixed population. This population has been subject to natural selection in response to environmental stresses, such as harsh climate, low-quality feeds, poor management, and strong disease challenge. Here, we combine two complementary sets of analyses, genome-wide association (GWA) and signatures of selection (SoS), to identify genomic regions that contribute to variation in milk yield and/or contribute to adaptation in admixed dairy cattle of Kenya. Our GWA separates SNP effects due to ancestral origin of alleles from effects due to within-population linkage disequilibrium. The results indicate that many genomic regions contributed to the high milk production potential of modern dairy breeds with no region having an exceptional effect. For SoS, we used two haplotype-based tests to compare haplotype length variation within admixed and between admixed and East African Shorthorn Zebu cattle populations. The integrated haplotype score (iHS) analysis identified 16 candidate regions for positive selection in the admixed cattle while the between population Rsb test detected 24 divergently selected regions in the admixed cattle compared to East African Shorthorn Zebu. We compare the results from GWA and SoS in an attempt to validate the most significant SoS results. Only four candidate regions for SoS intersect with GWA regions using a low stringency test. The identified SoS candidate regions harbored genes in several enriched annotation clusters and overlapped with previously found QTLs and associations for different traits in cattle. If validated, the GWA and SoS results indicate potential for SNP-based genomic selection for genetic improvement of smallholder crossbred cattle.
Keywords: GWAS; admixed cattle; haplotype; local ancestry inference; signatures of selection.
Copyright © 2020 Aliloo, Mrode, Okeyo and Gibson.
Figures

References
-
- Ariffin J. K., Kapetanovic R., Schaale K., Gatica-Andrades M., Blumenthal A., Schroder K., et al. (2016). The E3 ubiquitin ligase RNF144B is LPS-inducible in human, but not mouse, macrophages and promotes inducible IL-1β expression. J. Leukocyte Biol. 100, 155–161. 10.1189/jlb.2AB0815-339R - DOI - PubMed
LinkOut - more resources
Full Text Sources

