Impact of Diverse Data Sources on Computational Phenotyping
- PMID: 32582289
- PMCID: PMC7283539
- DOI: 10.3389/fgene.2020.00556
Impact of Diverse Data Sources on Computational Phenotyping
Abstract
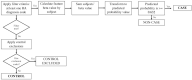
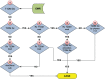
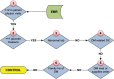
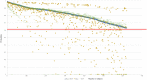
Electronic health records (EHRs) are widely adopted with a great potential to serve as a rich, integrated source of phenotype information. Computational phenotyping, which extracts phenotypes from EHR data automatically, can accelerate the adoption and utilization of phenotype-driven efforts to advance scientific discovery and improve healthcare delivery. A list of computational phenotyping algorithms has been published but data fragmentation, i.e., incomplete data within one single data source, has been raised as an inherent limitation of computational phenotyping. In this study, we investigated the impact of diverse data sources on two published computational phenotyping algorithms, rheumatoid arthritis (RA) and type 2 diabetes mellitus (T2DM), using Mayo EHRs and Rochester Epidemiology Project (REP) which links medical records from multiple health care systems. Results showed that both RA (less prevalent) and T2DM (more prevalent) case selections were markedly impacted by data fragmentation, with positive predictive value (PPV) of 91.4 and 92.4%, false-negative rate (FNR) of 26.6 and 14% in Mayo data, respectively, PPV of 97.2 and 98.3%, FNR of 5.2 and 3.3% in REP. T2DM controls also contain biases, with PPV of 91.2% and FNR of 1.2% for Mayo. We further elaborated underlying reasons impacting the performance.
Keywords: computational phenotyping; diverse data sources; phenotyping algorithms; rheumatoid arthritis; type 2 diabetes mellitus.
Copyright © 2020 Wang, Olson, Bielinski, St. Sauver, Fu, He, Cicek, Hathcock, Cerhan and Liu.
Figures

References
-
- Aletaha D., Neogi T., Silman A. J., Funovits J., Felson D. T., Bingham C. O., et al. (2010). 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthr. Rheum 62 2569–2581. 10.1002/art.27584 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

