Extracellular Vesicles of Human Periodontal Ligament Stem Cells Contain MicroRNAs Associated to Proto-Oncogenes: Implications in Cytokinesis
- PMID: 32582296
- PMCID: PMC7287171
- DOI: 10.3389/fgene.2020.00582
Extracellular Vesicles of Human Periodontal Ligament Stem Cells Contain MicroRNAs Associated to Proto-Oncogenes: Implications in Cytokinesis
Abstract
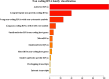
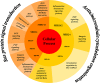
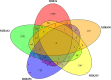
The human Periodontal Ligament Stem Cells (hPDLSCs) exhibit self-renewal capacity and clonogenicity potential. The Extracellular Vesicles (EVs) secreted by hPDLSCs are particles containing lipids, proteins, mRNAs, and non-coding RNAs, among which microRNAs, that are important in intercellular communication. The purpose of this study was the analysis of the non-coding RNAs contained in the EVs derived from hPDLSCs using Next Generation Sequencing. Moreover, our data were enriched using bioinformatic tools. The analysis highlighted the presence of non-coding RNAs and five microRNAs: MIR24-2, MIR142, MIR335, MIR490, and MIR296. Our results show that these miRNAs target the genes classified in two terms of the Gene Ontology: "Ras protein signal transduction" and "Actin/microtubule cytoskeleton organization." Noteworthy, the in-deep analysis of our EVs highlights that the miRNAs could be implicated in the silencing of proto-oncogenes involved in 12 different types of tumors.
Keywords: cytokinesis; extracellular vesicles; human periodontal ligament stem cells; microRNAs; next generation sequencing; non-coding RNAs.
Copyright © 2020 Chiricosta, Silvestro, Gugliandolo, Marconi, Pizzicannella, Bramanti, Trubiani and Mazzon.
Figures
References
-
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 281–297. - PubMed
LinkOut - more resources
Full Text Sources

