Epstein-Barr virus BART microRNAs in EBV- associated Hodgkin lymphoma and gastric cancer
- PMID: 32582365
- PMCID: PMC7310352
- DOI: 10.1186/s13027-020-00307-6
Epstein-Barr virus BART microRNAs in EBV- associated Hodgkin lymphoma and gastric cancer
Abstract
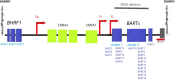
Background: EBV produces miRNAs with important functions in cancer growth, tumor invasion and host immune surveillance. The discovery of EBV miR-BARTs is recent, and most of their functions are still unknown. Nonetheless, some new studies underline their key roles in EBV-associated malignancies.
Main body: In EBV-associated tumors, the expression profile of miR-BARTs varies according to the cell type, autophagic process and signals received from the tumor microenvironment. By the same way of interest is the interaction between tumor cells and the tumor environment by the release of selected EBV miR-BARTs in addition to the tumor proteins trough tumor exosomes.
Conclusion: In this review, we discuss new findings regarding EBV miR-BARTs in Hodgkin lymphoma and gastric cancer. The recent discovery that miRNAs are released by exosomes, including miR-BARTs, highlights the importance of tumor and microenvironment interplay with more specific effects on the host immune response.
Keywords: BamHI fragment a rightward transcript; EBV; Epstein-Barr virus; miRNA; Gastric carcinoma cancer; HL; Hodgkin lymphoma; GC; Micro RNA; BART.
© The Author(s) 2020.
Conflict of interest statement
Competing interestsThere are no conflicts of interest in connection with this paper, and the material described is not under publication or consideration for publication elsewhere.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

