Detection of Alpha, Beta, Gamma, and Unclassified Human Papillomaviruses in Cervical Cancer Samples From Mexican Women
- PMID: 32582561
- PMCID: PMC7296070
- DOI: 10.3389/fcimb.2020.00234
Detection of Alpha, Beta, Gamma, and Unclassified Human Papillomaviruses in Cervical Cancer Samples From Mexican Women
Abstract
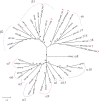
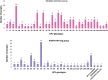
Background: Cervical cancer (CC) is associated to high-risk human papillomavirus (HPV) infections, for this reason it is crucial to have sensitive and accurate HPV diagnostic tests. To date, most research is focused on HPVs within the Alphapapillomavirus (α-PVs) genus and little attention has been paid to cervical infections with other HPV genotypes, like those of the Betapapillomavirus (β-PVs) and Gammapapillomavirus (γ-PVs) genera. The aim of this study was to determine the HPV genotypes from different genera in women with CC using Next-Generation Sequencing (NGS). Methods: The study comprised 48 HPV positive CC samples evaluated with the Linear Array HPV Genotyping test and individually sequenced by 454 NGS using PGMY09/11 and FAP primers. To determine the HPV genotypes present in each sample, the obtained sequences were compared with all HPV L1 gene reference sequences from the Papillomavirus Episteme database (PaVE). Moreover, 50 HPV positive low-grade cervical lesion samples individually genotyped with NGS were also included to determine the genotypes present preferentially in CC patients. Results: Among the 48 CC samples, 68.75% consisted of multiple HPV infections, 51 different genotypes were detected, of which 7 are still unclassified, 28 belong to α-PVs (6, 11, 16, 18, 26, 30, 33, 35, 39, 42, 43, 44, 45, 51, 52, 53, 54, 59, 62, 66, 68, 69, 70, 71, 74, 81, 102, 114), 10 to β-PVs (5, 12, 21, 37, 38b, 47, 80, 107, 118, 122), and 6 to γ-PVs (101, 103, 123, 135, 147, 214). Among them, HPV16 was the most prevalent genotype (54.2%), followed by HPV18 (16.7%), HPV38b (14.6%), and HPVs 52/62/80 (8.3%). Some genotypes were exclusively found in CC when compared with Cervical Intraepithelial Neoplasia grade 1 (CIN1) samples, such as HPVs 5, 18, 38b, 107, 122, FA39, FA116, mSK_120, and mSK_136. Conclusions: This work demonstrates the great diversity of HPV genotypes detected by combining PGMY and FAP primers with NGS in cervical swabs. The relatively high attribution of β- and γ- PVs in CC samples suggest their possible role as carcinogenic cofactors, but deeper studies need to be performed to determine if they have transforming properties and the significance of HPV-coinfections.
Keywords: HPV; Mexico; alpha; beta; cervical cancer; gamma; papillomavirus.
Copyright © 2020 Flores-Miramontes, Olszewski, Artaza-Irigaray, Willemsen, Bravo, Vallejo-Ruiz, Leal-Herrera, Piña-Sánchez, Molina-Pineda, Cantón-Romero, Martínez-Silva, Jave-Suárez and Aguilar-Lemarroy.
Figures



References
-
- Aguilar-Lemarroy A., Vallejo-Ruiz V., Cortés-Gutiérrez E. I., Salgado-Bernabé M. E., Ramos-González N. P., Ortega-Cervantes L., et al. (2015). Human papillomavirus infections in Mexican women with normal cytology, precancerous lesions, and cervical cancer: type-specific prevalence and HPV coinfections. J. Med. Virol. 87, 871–884. 10.1002/jmv.24099 - DOI - PubMed
-
- Artaza-Irigaray C., Flores-Miramontes M. G., Olszewski D., Vallejo-Ruiz V., Limon-Toledo L. P., Sanchez-Roque C., et al. (2019). Cross-hybridization between HPV genotypes in the linear array genotyping test confirmed by next-generation sequencing. Diagn. Pathol. 14:31. 10.1186/s13000-019-0808-2 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

